15 Best Free Bioinformatics Tools for Genomic Testing 2022
Popular applications for bioinformatics are best for sequence analysis and curations. The best solutions in the field have key inbuilt computational and big data analysis tools for genome sequencing. Let us have a look at what else these applications are comprised of in the following list.
List of 15 Best Free Bioinformatics Tools for Genomic Testing 2022
(1) geWorkbench
Summary of PhD Program: geWorkbench is an open-source bioinformatics platform that offers a comprehensive and extensible collection of tools for the management, analysis, visualization, and annotation of biomedical data. Many kinds of analysis are supported. For microarrays, there are tools for filtering and normalization, basic statistical analyses, clustering, network reverse engineering, as well as many common visualization tools.
(2) BioPerl
BioPerl is a collection of Perl modules that facilitate the development of Perl scripts for bioinformatics applications. It has played an integral role in the Human Genome Project.
(3) Unipro UGENE - Integrated Bioinformatics Tools
UGENE is free open-source cross-platform bioinformatics software It works perfectly on Windows, macOS and Linux and requires only a few clicks to install.
(4) BioJava
BioJava will run on any computer with a Java virtual machine complying to the Java specifications. Java implementations for Linux, Windows, and Solaris are available to download from Oracle’s java website. Recent versions of MacOS X include a suitable Java implementation as standard. Java is also available on many other platforms: if in doubt, contact your vendor. BioJava binaries are distributed in .jar (Java ARchive) format.
(5) Biopython
Biopython is a set of freely available tools for biological computation written in Python by an international team of developers. It is a distributed collaborative effort to develop Python libraries and applications which address the needs of current and future work in bioinformatics. The source code is made available under the Biopython License, which is extremely liberal and compatible with almost every license in the world.
(6) InterMine
InterMine integrates heterogenous data sources, making it easy to query and analyse data. InterMine’s tables allow you to easily drill down into data. It’s easy to filter data, add additional columns, navigate to report pages.
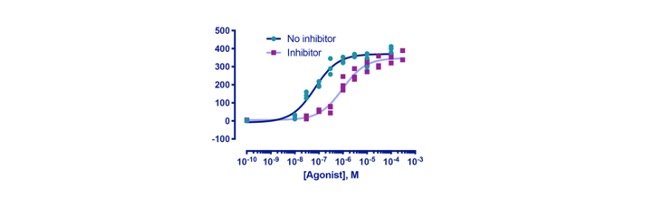
(7) Integrative Genomics Viewer (IGV)
The Integrative Genomics Viewer (IGV) is a high-performance, easy-to-use, interactive tool for the visual exploration of genomic data. It supports flexible integration of all the common types of genomic data and metadata, investigator-generated or publicly available, loaded from local or cloud sources.
(8) GROMACS
GROMACS is a versatile package to perform molecular dynamics, i.e. simulate the Newtonian equations of motion for systems with hundreds to millions of particles. It is primarily designed for biochemical molecules like proteins, lipids and nucleic acids that have a lot of complicated bonded interactions, but since GROMACS is extremely fast at calculating the nonbonded interactions (that usually dominate simulations) many groups are also using it for research on non-biological systems, e.g. polymers.
(9) Taverna Workbench Bioinformatics 2.5
Taverna Workbench Bioinformatics is an edition of Taverna Workbench that includes support for building and executing bioinformatics workflows using bioinformatics data and analytical services such as BioMart and BioMoby.
(10) EMBOSS
EMBOSS is “The European Molecular Biology Open Software Suite”. EMBOSS is a free Open Source software analysis package specially developed for the needs of the molecular biology (e.g. EMBnet) user community. The software automatically copes with data in a variety of formats and even allows transparent retrieval of sequence data from the web. Also, as extensive libraries are provided with the package, it is a platform to allow other scientists to develop and release software in true open source spirit. EMBOSS also integrates a range of currently available packages and tools for sequence analysis into a seamless whole. EMBOSS breaks the historical trend towards commercial software packages.
(11) Clustal Omega
Clustal Omega is the latest addition to the Clustal family. It offers a significant increase in scalability over previous versions, allowing hundreds of thousands of sequences to be aligned in only a few hours. It will also make use of multiple processors, where present. In addition, the quality of alignments is superior to previous versions, as measured by a range of popular benchmarks.
(12) BLAST
The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify members of gene families.
(13) bedtools
Collectively, the bedtools utilities are a swiss-army knife of tools for a wide-range of genomics analysis tasks. The most widely-used tools enable genome arithmetic: that is, set theory on the genome. For example, bedtools allows one to intersect, merge, count, complement, and shuffle genomic intervals from multiple files in widely-used genomic file formats such as BAM, BED, GFF/GTF, VCF. While each individual tool is designed to do a relatively simple task (e.g., intersect two interval files), quite sophisticated analyses can be conducted by combining multiple bedtools operations on the UNIX command line.
(14) Bioclipse Open Source Bioinformatics Tool
Bioclipse is a rich client, which means it is a downloadable application that run on your local computer but also gives the possibility to communicate with servers for data retrieval and computational services. The powerful plugin architecture is based on Eclipse, and results in a responsive, integrated user interface designed for simple and intuitive operations that at the same time is easy to extend with custom functionality.
(15) Bioconductor
The mission of the Bioconductor project is to develop, support, and disseminate free open source software that facilitates rigorous and reproducible analysis of data from current and emerging biological assays. We are dedicated to building a diverse, collaborative, and welcoming community of developers and data scientists.
Share