Category: Biology
-
Exons: Definition, Function, and Example
Continue ReadingExons, Coding Sequence, and Genes
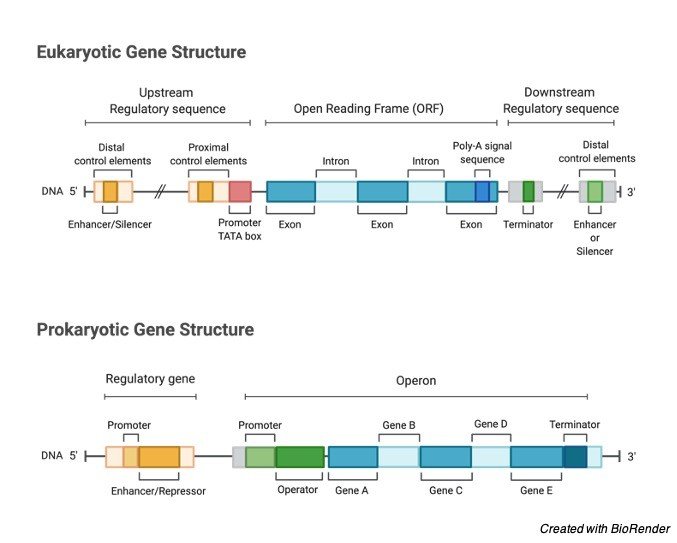
Exon is a part or a portion of a gene which codes for a particular amino- acids. In the plant and animal cells, most of the gene sequences are broken by one or more sequences of DNA, and they are named as Introns.
These parts of the gene sequences which are expressed as proteins are called as Exons.
The name is because, exons express, and introns do not express in terms of proteins, as introns come in between or to the interfere with the exons.
Exons are a part of the RNA, which codes for the proteins. when RNA gets transcribed initially it seems to be very large piece of molecule.
It is also very important to known that the RNAs here are the exons. These are the large chunks of the RNA which gets out excised. It is also important to know that exons are said to be excised because they move away.
The exons are the ones which stay in the mature RNA, and codes for the amino acids that are necessary.
What are Exons?
Exon is the particular part of a gene which codes a specific part of a mature RNA, which have been produced by the gene after removing the introns by splicing the RNA.
The term Exon is referred to both DNA sequencing that is present within the genome and the respective sequences that is present in the transcripts of RNA.
During RNA splicing introns are removed and the exons are let to join covalently along with-it ad a part of mature messenger RNA.
As the entire set of genes constitute a genome, the whole set of exons form an exome.
The term exon has been derived due to its capability of expressing the region of the gene.
It was termed by an American biochemist named Walter Gilbert in the year 1978. Where the notion of cistron was replaced by the unit of transcription containing regions that will be lost from the mature messenger RNA, and are called as introns. And the one which is expressed is known as Extron’s.
Contributions of Exons
Even the unicellular eukaryotes such as the yeast, doesn’t have introns or any other metazoans, vertebrate genomes have a large ratio of the non-coding DNA.
For instance if a human genome contains 1.1 percentage of the exons, there will be about 24 percentage of introns along with the remaining 75 percentage as the genome which will be as intergenic DNA.
All the eukaryotic genes in a gene bank have an average of 5.48 exons per gene. This exon encoded more than 30 – 36 amino acids.
The longest exon that is present in the human genome is 11,555 base pairs long, several exons are found just in 2 base pair long genome.
A single nucleotide of exon has been reported from the Arabidopsis genome.
Structure and Function of Exons
Exon is a precursor of messenger RNA. Exon includes both sequences which codes for amino acids and untranslated sequences.
In genes which code for proteins has the exons which includes both the protein coding sequence and the translational regions from 5’ to 3’ end.
Always the first exon includes both of the sequences, i.e., 5’UTR and the first part of the coding sequence, but exons which are containing only the regions of 5’UTR occurs in some genes, which is said that UTR contains introns.
Some of the non-coding transcripts of RNA has both exons and introns. Mature mRNA which originates from the same gene need not include the same exons, because introns present in the pre-mRNA can be removed by using the process of alternative splicing.
Exonization is the creation of the new exon, which is results as the mutation in the introns.
Exon Trapping
Exon trapping or gene trapping is a technique of molecular biology which exploits an existence of the intron-exon splicing in order to find the new genes.
Initially the trapped gene exon is spliced into an exon which is contained in the insertional DNA.
The new exon forms a ORF for a reporter gene which can express using the enhancers which is able to control the target gene.
Researches found that a new gene have been trapped at the time of expressing the reporter gene.
Splicing may be modified experimentally so that the targeted exons were excluded from the mature mRNA which transcripts by blocking the activity of the splice-directing small nuclear ribonucleoprotein particles to the pre-mRNA using the Morpholino antisense oligos, it has become the stranded technique in the field of developmental biology.
Where as the Morpholino oligos be targeted he prevent the molecules which regulates the splicing of enhancers, and suppressors from binding to pre-mRNA by altering the patterns.
Exon Shuffling
Exon shuffling is a one of the molecular mechanisms which is used for the formation of new genes.
It is one of the process through which two or more exons from the different genes are brought together ectopically, or the exon may be duplicated to create a new exon intron structure.
This technique for first introduced by Walter Gilbert in the year 1978. Where as the existence of introns could play an important role in evolution of proteins.
It has also been found that recombination within the intron can help in assorting the exons independently.
The respective segmental which are present in the middle of the introns can create hotspots for the recombination to shuffle the sequence of exons, which is known as exonic sequences.
Exon Citations
Share
Similar Post:
-

Introns: Definition, Function, and Example
Continue ReadingIntrons, Non-Coding, and Genes
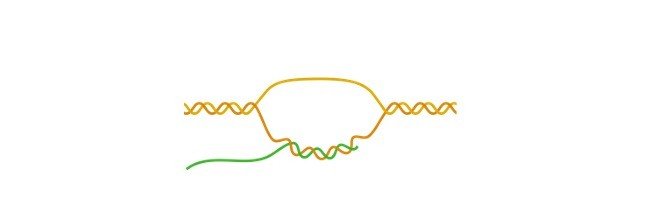
In some cases, not all the genes in the sequence of DNA are used to make a protein. Introns are the non-coding segments of the RNA which transcripts or encodes the DNA, which are spliced out before RNA molecule is being translated into the protein.
The sections of RNA or DNA which codes for proteins are known as exons. Following transcription, the new strands of immature m-RNA (messenger RNA) are produced, which is known as pre-mRNA, that contains both exons and introns.
The pre mRNA molecule under goes few modification processes in the nucleus of the cell which is known as splicing.
During splicing the non-coding introns are spliced out and only the coding exons are left over.
Through this process of splicing many messenger RNA are translated into a protein. Introns are also known as intervening sequences.
What are Introns?
Introns is defined as any of the nucleotide sequence within a gene which is removed by splicing the RNA during maturation of the final product of RNA.
In other words, it can also be said that, introns are the non-coding regions of a RNA transcript, or the DNA encoding it; which are eliminated by splicing before the process of translation.

The term intron has been derived from the name intragenic region, which means that it is a region present inside a gene.
It also means that intron refers to both the DNA sequence inside a gene and the corresponding sequence within an RNA transcript.
Sequences are joined together in final mature RNA after RNA splicing as exons.
Introns are present in the genes of almost all organisms including many of the viruses and may be located in a wide range of the genes, including those which produces proteins, ribosomal RNAs and the transfer RNAs.
When these proteins are generated from an intron containing genes, RNA splicing takes place as a part of the processing of RNA pathway which follows transcription and procedes translation.
Discovery of Introns
Introns were first observed in the protein-coding regions of the adenovirus and they were subsequently identified in the genes; encoding the transfer RNA and the ribosomal RNA.
Introns are found to be present in a wide variety of genes throughout the organisms within all the biological kingdoms.
The concept that genes are spliced by the introns was discovered by the scientists Phillip Allen Sharp and Richard Roberts in the year 1977.
The term intron was introduced by an American biochemist Walter Gilbert.
From the term Intron itself it is known that intron is something which refers to the intervening of the sequences which refers to any of the several families in the internal nucleic acid sequences which are not present in the final gene product, including the introns, untranslated regions and the nucleotides that are removed by editing the RNA in addition to that of the introns.
Classification of Introns
Splicing of all the introns contains RNA molecules which is exactly similar. Many types of introns are identified by examining the structure of intron by sequencing DNA analysis, along with the genetic and biochemical analysis of RNA during splicing.
Minimum four distinct classes of introns are identified. Introns in nuclear protein coding genes are removed by spliceosomes known as splicesomal introns.
Introns present in a nuclear and archaeal transfer RNA genes which are removed by the proteins.
Self-splicing group I introns are removed by catalyzing the RNA. Self-splicing group II introns are removed by the catalyzing of RNA.
Where as the Group III introns are proposed as a fifth family, which mediates their splicing. This is similar to the group II introns and also possible with the spliceosome introns.
I. Spliceosomal Introns
Nuclear pre-mRNA introns are generally classified by the specific sequence of introns which are located at the boundaries that are between introns and exons.
These sequences are identified by the spliceosome molecules of RNA during the time of initiation of splicing reactions.
It also contains a branch point, a specific point for sequencing a nucleotide near a 3’end of the intron during the process of splicing which generates a branched intron.
Apart from these three short conserved elements, nuclear pre- m RNA intron sequences are considered as a highly variable it also contains longer surrounding Extron’s.
II. tRNA Introns
t-RNA introns which are referred to as transfer RNA introns depends upon proteins for the removal of a specific location within the anticodon loop of an unsliced tRNA precursors and they are removed by an enzyme tRNA splicing endonuclease.
The exons are then linked together by a second protein by tRNA splicing ligase. It is also said that self-splicing introns are found in tRNA genes in few cases.
III. Group 1 & 2 Introns
These introns are commonly found in genes which encodes proteins such as m-RNA, t-RNA and r-RNA in a wide range of living organisms. After transcription in RNA, group I and group II introns make an extensive internal interaction which allows them to fold into a specific and a complex three-dimensional structure.
This complex structure allows few of the group I and group II introns to self-splice, which means that introns containing the molecules of RNA can rearrange its own co-valent structures to precisely remove the intron and there by it can link the exons together in an appropriate order.
Where as in few cases some other intron binding proteins are involved for splicing which assist in folding the intron in a three-dimensional way which is necessary for the activity of splicing.
However, the introns of group I and group II are differentiated by a specific set of internally conserved sequences and folded structures.
Where group I introns contains non-encoding guanine nucleotide to initiate the splicing by adding it to the 5’end of the excised intron.
The group II intron are differentiated by different set of internal conserved sequences where the group II molecules contain introns that are generated by branching introns.
Functions of Introns
Introns does not encode the protein products, where as they are integral to the expression of the gene for regulation.
Whereas some introns themselves encodes the functional RNAs by processing after the splicing process to generate the molecules of non-coding RNA.
Alternative splicing is widely used in generating the multiple proteins from the single gene.
Further few introns play an important role in expressing the wide range of gene regulating functions as nonsense-mediated decay and the mRNA export.
Introns Citations
Share
Similar Post:
-

DNA Template Strand: Definition, Types, and Functions
Continue ReadingWhat is DNA Template Strand?
A DNA template strand generally refers to the strand which is used by the enzyme DNA polymerases and RNA polymerases to attach with the complementary bases during the process of replication of DNA or at the time of transcription of RNA respectively.
In such cases, wither the molecule moves down towards the strand in the direction of 3’ to 5’ end and at each of the subsequent bases, it adds a complement to the current base of the DNA to the growth of nucleic acid strand which is being created by the 5’ to 3’ end.
The new strand created by the complementary, it matches the opposing strands of the DNA, which is known as coding strand.
Features of DNA Template Strand
Template strand in the DNA is also known as anti-sense strand; which is one of strand in the DNA that is present after they are being unwounded by the enzyme DNA helicase while the process of transcription.
Ribonucleotide triphosphates align themselves along the anti-sense strands by the base pairing made by Watson-crick.
In eukaryotes the ribonucleotides are further joined by the RNA polymerases II. Where as in prokaryotes the pre-mRNA, which is complementary to the template strand.
Transcription ends when the RNA polymerases reach the stop coding. In eukaryotes splicing takes place in order to remove the non-coding regions of the DNA to gives the final mRNA strand.
Then further the mRNA strand leaves the nucleus through the nuclear pore and is then translated by the ribosome, which is one of the cell organelles to form a sequence of amino acids Ans forms a necessary protein.
Characteristic DNA Template Strand
Template strand consists of an anticodon. It contains the same nucleotide sequences as such of the tRNA. Where the codon and template strand are the two different strands of the double strand DNA.
The template strand acts as a base for transcribing the mRNA and the others determines the appropriate base sequence for the mRNA.
It actually acts as a template for the synthesis of RNA. It moves in a direction of 3’ to 5’.
The RNA polymerases read the template strand in the direction of 3’ – 5’ end. It has a nucleotide base sequence which is complementary to both the coding strands and also to the mRNA.
During genetic coding the template strands have the anticodons. During the formation of hydrogen bonds, the bonds formed in the template strands are temporary and whereas the newly synthesised mRNA occurs at the time of transcription.
Function of DNA Template Strand
As said before template strand is one of the DNA strands whose sequence of bases helps in building up the mRNA through the complementary base sequencing.
Template strand which is also known as antisense strands runs in the direction of 3’ to 5’ ends, which runs opposite to the coding strands.
Template strand contains the complementary nucleotide sequences which are further transcribes into the m-RNA.
After the process of transcription, the mRNA is converted into the mature mRNA, which undergoes specific post-transcriptional modifications.
The template strand also contains the anticodons which carries the triplet codes or the triplet nucleotide sequences complementary to anticodon sequence of a t-RNA.
The anticodon thus helps in attaching the specific amino acid to the t-RNA and forms proteins or a polypeptide chain through the assistance of the r-RNA.
An enzyme RNA polymerase reads the template strand to synthesis the RNA transcript by recognising the specific sequences.
Hence, RNA polymerase is considered as the one which decides the initiation of the transcription and also in the termination of the translation process.
Coding DNA Template Strand
Template and coding strands are the terms generally used to describe the strands which are present in the DNA. During the process of transcription, one of the two strands in the double stranded DNA serves as a template strand.
This template strand walks in the direction of 3’ to 5’ end. Where as the other strand which is present in the DNA, other than the template strand is known as coding strand.
Template strand is responsible for the sequencing amino acid for synthezing the polypeptide chain.
The main difference to be considered between the coding and template strand is that the template strand serves as the template for the transcription where the coding strand contains the exact and the same sequence of the nucleotides in mRNA, expect the nucleotide thymine.
DNA Template Strand and Transcription
A single strand of RNA is synthesised using a double stranded DNA molecule as the template. The two strands of the molecule of DNA are separated from one another by exposing the nitrogenous bases.
Only one of the strands is actively used as a template in the process of transcription. The strand which is used as a template is also known as template strand or sense strand.
The complementary strand of the DNA is the one which is not used and is called as the nonsense strand or the antisense strand.
The RNA sequence which is made up of a direct copy of the nitrogenous bases in the template strand.
If Guanine base is a part of sequence on the template DNA strand, then the molecule of RNA has a Cytosine base which is added to its sequence at that point.
In the molecule of RNA, uracil substitutes for Thymine.
DNA Template Strand Citations
- Probing DNA polymerase-DNA interactions: examining the template strand in exonuclease complexes using 2-aminopurine fluorescence and acrylamide quenching. Biochemistry . 2007 Jun 5;46(22):6559-69.
- Detection of template strand switching during initiation and termination of DNA replication of porcine circovirus. J Virol . 2004 Apr;78(8):4268-77.
- DNA template-assisted inhibition of tyrosinase activity. Int J Biol Macromol . 2015 Aug;79:278-83.
Share
Similar Post:
-
Transcription: Definition, Mechanism, Steps, and Functions
Continue ReadingWhat is Transcription?
In general, we all have transcribed something, such as messages or voice mails by writing in a paper or coping in something else. Like wise transcription is nothing but a kind of process in which the information in the molecule is rewritten, so it can be said that transcription is something that we all follow in our daily life, in the same way our cells also has some way of transcribing the proteins and the hereditary information.
Transcription in our cells is carried in a specialised and a narrow manner. However, in biology transcription is defined as the process of copying the information in our DNA, into a sequence of a gene as the component of the RNA.
Transcription is considered as the first step in expression of the gene where the information from a gene is constructed as a functional product in the form of proteins.
The main aim of transcription is to make a copy of RNA, in a DNA sequence of the gene. Considering the protein coding gene, the copy of RNA which is transcript and thus carries an information needed for building a polypeptide which is known as protein or the subunits of protein.
In eukaryotes transcription needs to go through some of the steps of processing before translating into proteins.
Mechanism of Transcription?
Transcription is a process by which the information from one strand is copied to the new molecule which is present in the messenger RNA, which is most commonly called as mRNA.
DNA stably stores the genetic material safely in the nucleic acid of the cells as a template or a reference.
At that time, mRNA is comparable to copy from the reference cell as it carries the similar information which is not a different copy of the DNA segment as its sequence is complementary to the template of the DNA.

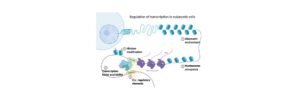
Transcription is usually carried out by an enzyme called as RNA polymerase and the number of accessory proteins which are called as Transcription factors.
These Transcription factors helps in binding the specific DNA sequences that are commonly called as enhancers and a promoter sequences, in order to recruit the RNA polymerases in an appropriate transcription site.
Transcription factors and the RNA polymerases forms a complex known as transcription initiation complex.
This complex initiate transcription and the RNA polymerases begins the synthesis of mRNA by matching the complementary bases of the original strand of DNA.
The mRNA molecule is an elongated. Once the strand is synthesised completely, transcription is terminated.
The newly formed mRNA copies of the gene serve as the blue prints for the synthesis of the protein during the process of translation.
Characteristic of Transcription
During the process of transcription, only one strand of the DNA is copied which is known as template strand and here the RNA molecules are single stranded and are called as messenger RNAs (mRNAs).
The DNA strand corresponds the mRNA which is known as coding or sense strand.
Eukaryotes are the organisms which possess a nucleus, the transcription taking part in eukaryotes consists of an initial product known as pre-mRNA, which is extensively edited through splicing before the production of mature mRNA and the translation is being read by ribosomes, which is one of the cell organelles which serves as a site for synthesis of proteins.
Transcription of any one gene takes place at the location of chromosomes in that particular gene, which is relatively a short segment of the chromosome.
The active transcription process in a gene depends on the need for the activity in that particular gene in a specific tissue or a cell at a given time.
Transcription in Prokaryotes
Prokaryotes are the organisms which lacks the nucleus. Many genes in prokaryotes expresses a signal called as operators which are also known as operons, when a specialised protein named repressors bind to the DNA in an upstream which is a start point of transcription to prevent the access of the DNA with the help of RNA polymerase.
These repressor proteins prevent the transcription of the gene by physically blocking the action of the enzyme, RNA polymerase.
Typically, repressors are released from the action of blocking when receiving signals other molecules in the cell indicating that the genes have to be expressed.
Apart from some of the prokaryotes genes expresses signals to which activator proteins which bind to stimulate the transcription.
Transcription in Eukaryotes
Usually, the transcription in eukaryotes is more complicated than prokaryotes. Initially the RNA polymerase in higher organisms is a more complicated enzyme than a relative sub unit enzyme of prokaryotes.
In addition to this there are many accessory factors which helps in controlling the efficiency of the individual promoters.
These accessory proteins are generally called as transcription factors and it typically responds to the signals which arises within the cell that whether the transcription is required.
In many genes of humans many transcription factors are needed before proceeding the transcription which helps in efficient transcription.
Transcription factor either causes repression or activation of the gene while expressing in eukaryotes.
Transcription Steps
I. Initiation
This is the first step in the process of initiation, Here the RNA polymerase binds to the sequence of DNA, which is called as promoter and found near the beginning of the gene.
Where each gene has its own promoter. Once it is bound the RNA polymerases separates the strands of the DNA, which provides the template single stranded for the process of transcription.
II. Elongation
Here one strand of the template of the DNA, acts as a template for the enzyme, RNA polymerase. It reads this template in a base in an appropriate time.
The polymerase builds an RNA molecule out of complementary nucleotides, that makes a chain which grows from 5’ to 3’ end.
The transcript RNA carries the same information as the coding non template strand of the DNA, but it contains the Uracil base instead of the thymine(T).
III. Termination
Sequences are called as terminators signals where the RNA transcript is complete. Once they are transcribed, it causes the transcript to be released from the enzyme RNA polymerase.
One such example of a mechanism of termination is involving the formation of a hairpin the RNA.
Transcription Citations
Share
Similar Post:
-

RNA Polymerase: Function, Types, and Definition
Continue ReadingWhat is RNA Polymerase?
RNA polymerase plays an important role in carrying out the process of transcription. Transcription is a process where the genes of a DNA sequence is transcribed to make a molecule of RNA, where RNA polymerase plays an important role in this process.
Transcription begins when the enzyme RNA polymerase binds to the sequence of promoter beside the beginning of a gene sequencing either directly or through the helper proteins.
RNA polymerase uses one of the strands of the DNA, as a template; so, the strand of DNA is mentioned as template strand which makes a new molecule of complementary RNA and transcription ends in a process called termination.
Where termination depends on the sequence of the RNA, where the signal arises when the transcription is complete.
RNA polymerase is considered as one of the crucial enzymes as it carries out the process of transcription, where the process of copying the genetic material Deoxyribose ribonucleic acid into the Ribonucleic acid (RNA).
Because transcription is one of the most vital steps involved in using the information from genes in our DNA to make the essential proteins, Proteins act as key molecule which gives a cell structure and helps in running them continuously.
Generally, mushrooms have their lethal effect in producing the specific toxin, that attaches it to the crucial enzyme in our body, which is RNA polymerase.
Blocking the transcription process with the mushroom toxin causes many defects like liver failure and even death, because there will no RNA so no proteins will be produced. So, transcription is very much important in caring out a life process.
Role of RNA Polymerase in Transcription
RNA polymerases are the enzymes which transcribe the DNA into the RNA. Using the DNA template, this enzyme RNA polymerases builds a new molecule of RNA through base pairing.
For example, if there is any G i.e., Guanine in the DNA template, RNA polymerases act a C (cytosine) to the new developing strand of RNA.
RNA polymerases always build a new RNA strand in the direction of 5’ to 3’ end which helps in adding the nucleotides to the 3’ end of the strand.
RNA polymerases are usually the large enzymes which has multiple sub units even in the lower organisms like bacteria.
Where as the eukaryotes and humans have three variety of RNA polymerases, named as RNA Polymerase – I, II and III.
Each of this type has specialising transcribing factors in certain classes of genes.
Plants have additional polymerases apart from this as IV and V, which plays an important role in producing certain small RNAs.
RNA Polymerase in Initiating Transcription
To initiate the process of transcription by transcribing a gene, RNA polymerases act as a binding agent by binding to the DNA of the gene at the regions known as promoter.
Generally, this promoter conveys a message to the polymerase that where it has to be positioned on the DNA and where it has to start transcribing.
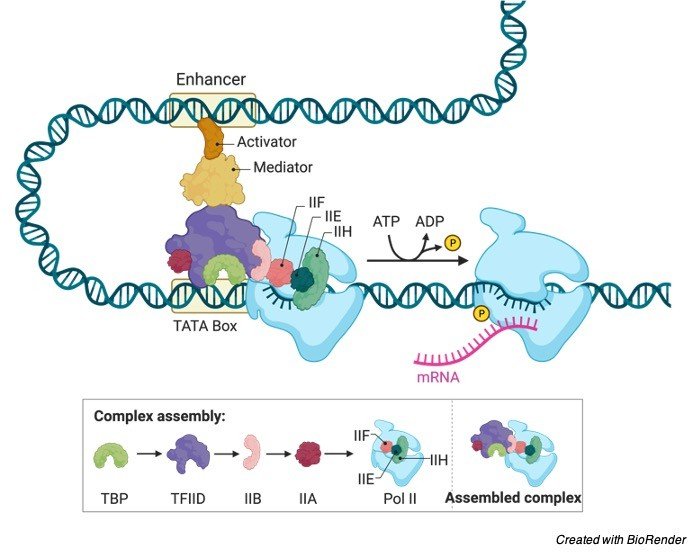
Each gene contains its own promoter, where the promoter contains the sequence of DNA which lets either the enzyme RNA Polymerase or its helper proteins to attach to the DNA. When the transcription bubble is formed, polymerase starts transcribing.
RNA Polymerase, Promoters, and Bacteria
Promoter in bacteria stands as a best example of how a polymerase works, Bacteria contains two types of important sequences of DNA, -10 and -35 elements.
RNA polymerases recognise and binds straitly to these sequences, where these sequences position the polymerase enzymes in the appropriate spot and initiates the transcribing in the target gene, and it also ensures the pointing that it acts in a right direction.
Once the enzyme RNA polymerase bounds up it opens the DNA and starts to function. Opening of DNA occurs at the -10 element, Here the strands can be separated easily because of the factors such as As and Ts, which helps in binding to each other with the hydrogen bonds.
The elements -10 and -35 derives its name from the nucleotides 35 and 10. The minus sign is denoted just to show that they are before the site of initiation.
RNA Polymerase, Promoters, and Humans
In eukaryotes like mammals and humans, the RNA polymerase in the cells does not directly attaches to promoters like that of the polymerases in the bacteria.
Instead of this helper proteins which are called as basal transcription factors which first binds to the promoters, and helps the RNA polymerases in the cells to get hold on to the DNA.
Several eukaryotic promoters consist of a sequence known as TATA box, which plays an important role similar to that of the -10 element.
It is actually recognised by the general transcription factors, which allows the other transcription factors and also the RNA polymerases to bind eventually.
It also contains a lot of As and Ts, which also makes it very easy to pull the strands of the part of DNA.
RNA Polymerase and Elongation
When the enzyme, RNA polymerase is positioned at the site of promoter, the next step of transcription begins, which is known as elongation.
Elongation is the stage where the strands of RNA get longer. During elongation these RNA polymerases moves along the single strand of DNA, which is commonly called as template strand in the direction of 3’ to 5’.
Each nucleotide in a template needs RNA polymerases to match it with a nucleotide of an RNA at a 3’ end in the RNA strand.
RNA Polymerase and Termination
RNA polymerases transcribe until the signals gets stopped. Ending the process of transcription is generally known as termination and it happens only when the polymerase transcribes one of sequence of DNA which is known as terminator.
RNA Polymerase Citations
Share
Similar Post:
-

CRISPR/Cas-9: Definition, Function, Mechanism, and Examples
Continue ReadingWhat is CRISPR/Cas-9?
The CRISPR/Cas-9 innovation has been a progressive invention in the study of quality adjustment. The innovation has given us an apparatus to change hereditary material in living life forms, by utilizing an instrument that was initially a safeguard framework in bacteria.
The CRISPR/Cas-9 framework comprises of two primary parts: the CRISPR quality and the Cas-9 protein.
CRISPR is an abbreviation for Clustered Regularly Interspaced Short Palindromic Repeats and are short, interspaced groupings of DNA, which are rehashes of one another.
In the middle of these indistinguishable strands of DNA are other more modest strands of DNA.
At the point when CRISPR was first found in E-Coli, specialists tracked down that the little successions in the middle of the rehashes were infection DNA.
This shows that when infection DNA assaults the cell, the bacteria has DNA of the infection put away or can store DNA.
The complex likewise holds the DNA for the nuclease-protein Cas. This protein can isolate a quality and cut it with sub-atomic scissors. Cas protein looks for the DNA, which it needs to cut, it opens up a hereditary succession by isolating it and afterward checks after the DNA that coordinates with its RNA.
If it matches Cas begins to annihilate the DNA by cutting it. On the off chance that it doesn’t coordinate, Cas shuts the DNA again and keeps looking for its coordinating with DNA.
History of CRISPR/Cas-9
Educator Jennifer Doudna and analyst Emmanuelle Charpentier fostered the CRISPR/Cas-9 framework in 2012.
They interpreted the safeguard framework in the bacteria Streptococcus pyogenes. From this framework they have made the ‘tracr-RNA-cr-RNA fabrication’ framework.
Basically, they trade the infection cr-RNA (CRISPR-RNA) with RNA of their decision. The tracr-RNA (tracer-RNA) holds the cr-RNA set up in the Cas-9 protein.
CRISPR/Cas-9 Technology
The framework comprises of the Cas-9 protein just as the cr-RNA-tracr-RNA, likewise, alluded to as the g-RNA (guide RNA). Besides, the framework likewise comprises of the host DNA that is embedded into the inactivated quality cut by Cas-9.
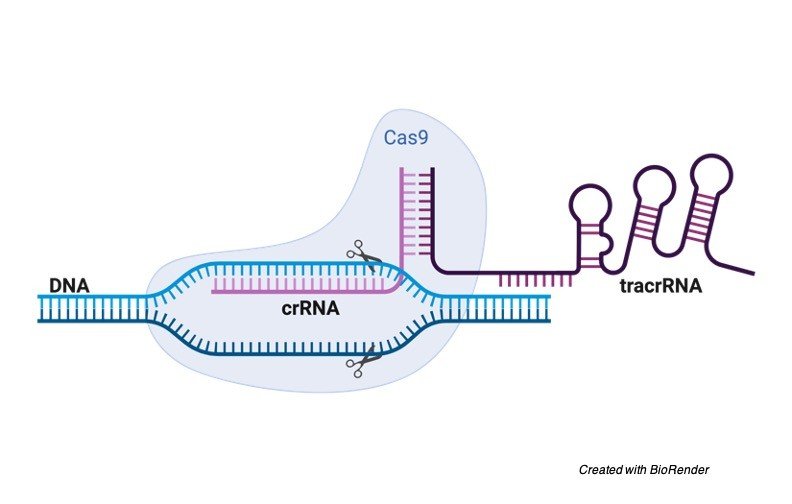
A g-RNA that codes for this specific DNA is created. The complex is embedded into a cell by utilizing a plasmid with the hereditary codes for the created g-RNA and the Cas-9 protein.
It is embedded into the Cas-9 protein. Cas-9 discovers the DNA that coordinates with its g-RNA and cuts it.
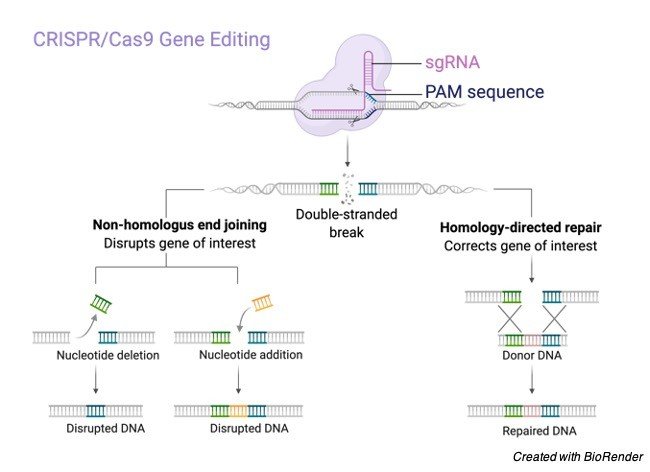
From here, you have a DNA grouping with a hole in it. Now and again the DNA will transform to fix the break, yet regularly it is feasible to inactivate the grouping and afterward embed the picked have DNA into the break and fixing it.
In this manner, it is feasible to adjust DNA arrangements by utilizing the CRISPR/Cas-9 technique and any creature can be effectively edited.Importance of CRISPR/Cas-9 Technology
The exactness of the CRISPR framework implies it very well may be utilized in various fields. In medication the instrument has been demonstrated to be incredibly valuable.
It has not been at this point endorsed for use on people for clinical preliminaries. Nonetheless, as indicated by Jennifer Doudna almost certainly, we will see supported clinical preliminaries and treatments on grown-up people inside the following ten years.
As indicated by Doudna, we can undoubtedly utilize CRISPR to potentially fix the hereditary imperfection that causes sickle cell anaemia.
There are as of now prospects in treating HIV with the technology also. CRISPR is widespread on all cell types, which opens up incalculable potential outcomes. It is being tried for use in microbiology-related agriculture also.
These employments of CRISPR are approaches to better our reality, which I consider to be a positive utilization of the innovation.
In any case, there are moral contemplations with regards to CRISPR by utilizing it prevalently without assent, for instance as to altering the genome in incipient organisms.
These are alterations, which change the ages of people, in this manner adjustments on a creature that influences its advancement.
A grown-up can offer agree to having something altered by utilizing the CRISPR technique, however an undeveloped organism can’t give its assent, and neither can its posterity that likewise gets these alterations.
Thusly, it makes it non-consensual and subsequently tricky. Most of the Danish Council on Ethics concurs that it is flippant to utilize CRISPR on embryos and concur. This is a part of the innovation that we ought not abuse.
Why CRISPR/Cas-9 Technology is Very Significant?
The CRISPR/Cas 9 procedure is one of various quality altering instruments. Many blessing the CRISPR/Cas9 strategy in view of its serious level of adaptability and precision in reordering DNA.
One reason for its prevalence is that it makes it conceivable to do hereditary designing on an extraordinary scale for an extremely minimal price.
How it varies from past hereditary designing procedures is that it takes into account the presentation or expulsion of more than each quality in turn.
This makes it conceivable to control a wide range of qualities in a cell line, plant or creature rapidly, diminishing the interaction from requiring various years to only weeks.
It is additionally extraordinary in that it isn’t species-explicit, so can be utilized on organic entities beforehand impervious to hereditary designing.
The method is as of now being investigated for a wide number of utilizations in fields going from horticulture through to human wellbeing.
In agribusiness it could help in the plan of new grains, roots and organic products. Inside the setting of wellbeing it could make ready to the improvement of new medicines for uncommon metabolic issues and hereditary illnesses going from hemophilia through to Huntingdon’s infection.
It is likewise being used in the making of transgenic creatures to deliver organs for transfers into human patients.
The innovation is likewise being examined for quality treatment. Such treatment expects to embed ordinary qualities into the cells of individuals who experience the ill effects of hereditary issues like cystic fibrosis, haemophilia or Tay Sachs.
A few new businesses have been established to misuse the innovation industrially and huge drug organizations are additionally investigating its utilization for drug revelation and improvement purposes.
The significance of the CRISPR/Cas9 was perceived with the granting of the Nobel Prize in Chemistry to Jennifer Doudna and Emmanuel Charpentier on seventh October 2020.
What is missed in the granting of the Prize is the huge job that numerous others, including Virginijus Siksnys, played in assisting with achieving the improvement of quality altering.
CRISPR/Cas-9 Citations
- The genome editing revolution: review. J Genet Eng Biotechnol . 2020 Oct 29;18(1):68.
- Harnessing CRISPR/Cas 9 System for manipulation of DNA virus genome. Rev Med Virol . 2019 Jan;29(1):e2009.
- CRISPR/Cas 9 genome editing and its applications in organoids. Am J Physiol Gastrointest Liver Physiol . 2017 Mar 1;312(3):G257-G265.
Share
Similar Post:
-
Single Nucleotide Polymorphism: Definition, Function, and Examples
Continue ReadingWhat is Single Nucleotide Polymorphism (SNP)?
A Single Nucleotide Polymorphism or SNP (articulated ‘clip’) is a little hereditary change, or variety, that can happen inside a DNA arrangement.
The four nucleotide A (adenine), C (cytosine), T (thymine), and G (guanine) determine the hereditary code. SNP variety happens when a single nucleotide, like A, replaces one of the other three nucleotide letters – C, G, or T.
By traditional meaning of polymorphism, the recurrence of the variety should be essentially 1% to qualify the nucleotide change as a polymorphism.
Those nucleotide changes that happens under 1% would be called uncommon variation. Since simply about 1.1 to 1.4% of an individual’s DNA groupings codes for proteins, most Single Nucleotide Polymorphism are found outside of coding successions.
Single Nucleotide Polymorphism lying outside the coding district regularly would not be relied upon to anily affect the aggregate of a creature.
Single Nucleotide Polymorphism found inside a coding grouping are specifically compelling to specialists as they are bound to adjust the natural capacity of a protein, albeit these progressions have substantially less uncommon impact than that of transformations.
Because of late advances in field of quality recognizable proof and portrayal, there has been a tremendous whirlwind of SNP revelation.
Discovering single nucleotide changes all through the human genome appears to be a mammoth work, yet, throughout the most recent 20 years, analysts have fostered various methods that makes it conceivable.
Every procedure utilizes a comparative non-indistinguishable technique to look at chosen locales of a DNA grouping acquired from numerous people who share a typical characteristic.
In each test, the outcome shows a distinction in the DNA tests when a SNP is identified in one individual in a pool under test.
Dissemination of Single Nucleotide Polymorphism
Single Nucleotide Polymorphism are not circulated consistently over the genome. A colossal number of Single Nucleotide Polymorphism are appropriated all through the non-coding district of the genome.
Since these districts are liberated from determination pressure, these progressions are chosen impartially and fixed over the long run.
The conveyance examples of the Single Nucleotide Polymorphism are variable even in a single chromosome.
For example, locales answerable for antigen show to the insusceptible framework, present on the chromosome 6, shows extremely high nucleotide inconstancy as opposed to different districts of a similar chromosome.
The Origin, Survival and Fixation of Single Nucleotide Polymorphism: The SNP is the primary wellspring of change in the genome and it represents 90% of all human polymorphism.
Types of Single Nucleotide Polymorphism
There are Two Types of Nucleotide Base Substitution;
Transition: Change, which represents almost 66% of all Single Nucleotide Polymorphism, happens between purines (for example A > G) or pyrimidines (for example C > T).
Transversion: Transversion happens among purines and pyrimidines (for example A > C and G > T).
Its Life can be Roughly Divided into 4 Phases:
1) Appearing by the method for point changes.
2) Surviving the determination pressing factor of the nature.
3) Spreading through ages.
4) Establishing itself essentially as 1% of all alleles.
The most continuous change in people is the transformation from CpG to TpG (a progress representing around 25% of all transformations).
This system causes decline in the quantity of CG dinucleotide in the genome since numerous in the long run becomes TG, while new CpG locales will be made by other less regular transformations.
Since simply 1.1% to 1.4% of the genome codes for proteins Single Nucleotide Polymorphism are probably going to happen at non-coding successions all the more regularly.
Regardless of whether the SNP happens at a coding succession, generally it may have an unobtrusive and non-harmful impact on the communicated proteins.
Changes representing pernicious impacts are ultimately eliminated from the genome by regular determination.
Consequently to accomplish the situation with a SNP, a point transformation ought to be non-injurious to be chosen.
Utilization of Single Nucleotide Polymorphism in Pharmacogenomics Studies
Reaction rates towards major and normal medications shift clearly among people. Single Nucleotide Polymorphism trait in a significant manner towards this wonder.
Utilizing Single Nucleotide Polymorphism to examine the hereditary qualities of medication reaction can possibly help in the formation of customized medication as clarified in.
As referenced before, Single Nucleotide Polymorphism may likewise be related with the digestion i.e., absorbance and leeway of remedial specialists.
As of now, there is no standard hereditary screening of medication using qualities to decide how a patient will react to a specific drug.
A treatment demonstrated powerful in one patient might be insufficient in others.
A few patients may likewise encounter unfavourable immunological response to a specific medication.
Thus, drug organizations limit their creation of medications for which an ‘normal’ patient will react. Subsequently a somewhat more modest gathering of patients holding onto any putative hereditary variety (for example a SNP), which renders them unfit to utilize that medication, stays untreated.
Numerous medications that may profit that little gathering of patients never make it to showcase as those medications would bring less benefit for the medication Industries.
Primary and Functional Importance of Single Nucleotide Polymorphism
Notwithstanding the Single Nucleotide Polymorphism happening in the coding succession of qualities, useful significance of Single Nucleotide Polymorphism has likewise been seen in non-coding DNA (for example introns) including administrative (for example advertisers, enhancers and so forth).
One genuine illustration of practical SNP is in a non-coding district is the tau quality. The design of tau exon 10 grafting administrative component RNA has been as of late interpreted and has been displayed to frame a stable collapsed stem-circle structure.
Single Nucleotide Polymorphism have generally been utilized to coordinate with a scientific DNA test to a suspect yet has been made old due to progressing STR-based DNA fingerprinting strategies.
Be that as it may, the improvement of cutting-edge sequencing (NGS) innovation may consider more freedoms for the utilization of Single Nucleotide Polymorphism in phenotypic pieces of information like identity, hair tone, and eye tone with a decent likelihood of a match.
This can moreover be applied to build the precision of facial reproductions by giving data that may somehow or another be obscure, and this data can be utilized to assist with distinguishing suspects even without a STR DNA profile match.
A few cons to utilizing Single Nucleotide Polymorphism versus STRs is that Single Nucleotide Polymorphism yield less data than STRs, and in this way more Single Nucleotide Polymorphism are required for examination before a profile of a suspect can be made.
Furthermore, Single Nucleotide Polymorphism intensely depend on the presence of an information base for near examination of tests.
Be that as it may, in examples with debased or little volume tests, SNP procedures are a superb option in contrast to STR techniques.
Single Nucleotide Polymorphism (instead of STRs) have a wealth of expected markers, can be completely computerized, and a potential decrease of required part length to under 100bp.
Single Nucleotide Polymorphism and Pharmacogenetics
A few Single Nucleotide Polymorphism are related with the digestion of various medications.
SNP’s can be transformations, like erasures, which can restrain or advance enzymatic action; such change in enzymatic action can prompt diminished paces of medication digestion.
The relationship of a wide scope of human illnesses like malignant growth, irresistible sicknesses (AIDS, infection, hepatitis, and so forth) immune system, neuropsychiatric and numerous different sicknesses with various Single Nucleotide Polymorphism can be made as important pharmacogenomic focuses for drug treatment.
Infection A solitary SNP may cause a Mendelian sickness, however for complex infections, Single Nucleotide Polymorphism don’t as a rule work independently, rather, they work in a joint effort with different Single Nucleotide Polymorphism to show an illness, for example, in Osteoporosis.
Perhaps the soonest achievement in this field was tracking down a solitary base transformation in the non-coding locale of the APOC3 (apolipoprotein C3 gene) that related with higher dangers of hypertriglyceridemia and atherosclerosis.
A few infections brought about by Single Nucleotide Polymorphism incorporate rheumatoid joint pain, crohn’s sickness, bosom malignancy, Alzheimer’s, and some immune system problems.
Huge scope affiliation contemplates have been performed to endeavours to find extra infection causing Single Nucleotide Polymorphism inside a populace, yet countless them are at this point unclear.
Single Nucleotide Polymorphism Citations
Share
Similar Post:
-

Genome Editing: Definition, Technology, and Examples
Continue ReadingGenome Editing: Good or Bad?
Jiankui Chinese specialist stunned mainstream researchers in 2018 in the wake of declaring he had effectively changed the genes of twin young ladies brought into the world in November to keep them from contracting HIV.
He had “secretly” coordinated an undertaking group that included unfamiliar staff and utilized “innovation of questionable security and adequacy” for unlawful human undeveloped organism gene-altering, specialists said.
However, such Genome Editingwork is prohibited in many nations, including China.
CRISPR-CAS9: a Wonderful Genome Editing Technology
CRISPR-CAS9 is an innovation that permits researchers to basically reorder DNA, raising any desire for genetic fixes for infection. Nonetheless, there are likewise worries about its security and morals.
CRISPR is a dynamic, flexible device that permits us to target almost any genomic area and conceivably fix broken genes.
It can eliminate, add or change explicit DNA arrangements in the genome of higher organic entities.
How does Genome Editing work?
Strange yet rehashed DNA structures that researchers had been noticing for quite a while were given a name. This name appointed was “Bunched consistently interspaced short palindromic rehashes” or CRISPR.
In 2012, researchers found that CRISPR is a critical piece of the “resistant framework”.
For example, when an infection enters a bacterium, it retaliates by cutting up the infection’s DNA. This kills the infection yet the bacterium stores a portion of the DNA.
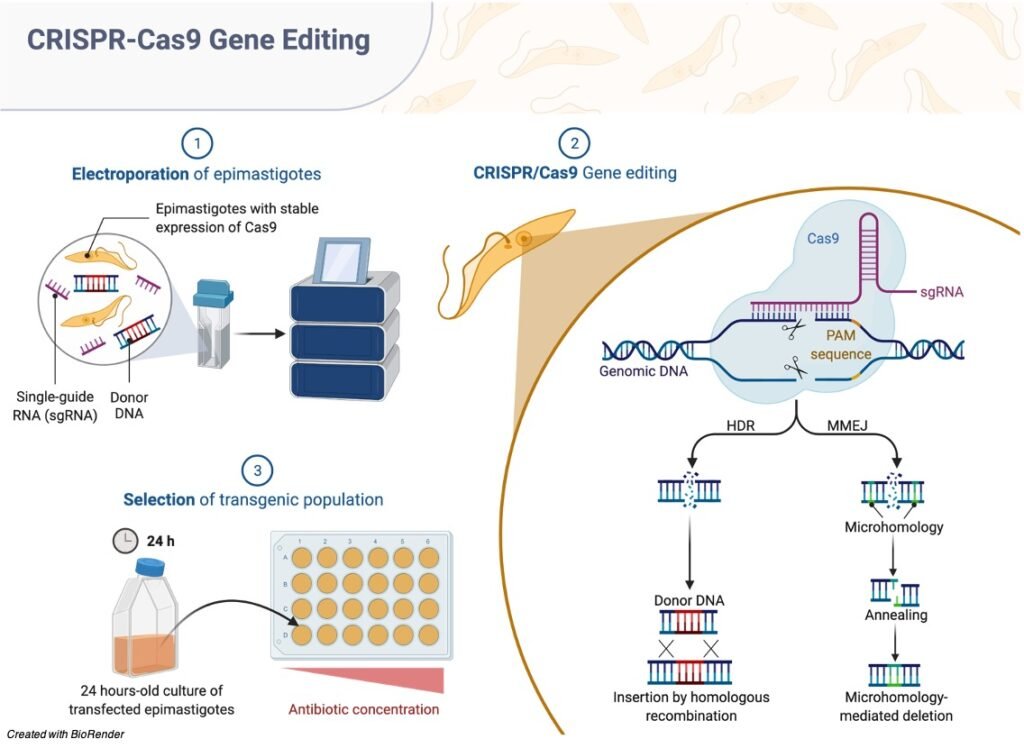
The following time there is an attack, the bacterium creates a protein called Cas9 which matches the put away fingerprints with that of the trespassers.
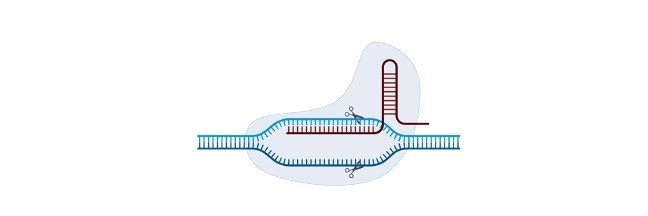
In the event that it matches, Cas9 can cut the attacking DNA. The CRISPR-Cas9 Genome Editingapparatus along these lines has two segments.
They are: a short RNA grouping that can tie to a particular objective of the DNA and the Cas9 chemical which goes about as sub-atomic scissors to cut the DNA.
To alter a gene of interest, the short RNA arrangement that consummately coordinates with the DNA grouping that needs to be altered is presented.
When it ties to the DNA, the Cas9 protein cuts the DNA at the designated area where the RNA grouping is bound.
When the DNA is cut, the normal DNA fix instrument is used to add or eliminate genetic material or make changes to the DNA.
Benefits of Genome Editing
CRISPR could be utilized to change sickness causing genes in undeveloped organisms brought to term, eliminating the flawed content from the genetic code of that individual’s future relatives too.
Genome altering (Genome Editing) might actually diminish, or even dispense with, the rate of numerous genuine genetic infections, lessening human experiencing around the world.
It may likewise be feasible to introduce genes that offer deep rooted insurance against disease.
CRISPR May Prove Useful in De-Extinction Efforts. For instance, Researchers are utilizing the incredible a Genome Editingdevice to reproduce the wooly mammoth.
CRISPR Could Create New, Healthier Foods: In horticultural yields, Crispr can possibly affect yield, infection obstruction, taste, and different characteristics.
Hardly any analyses have been finished. In the event that effective it can assist us with destroying the issue of yearning and lack of healthy sustenance.
Cons of Genome Editing
Rolling out irreversible improvements to each cell in the assortments of future youngsters and every one of their relatives would establish remarkably hazardous human experimentation.
There are issues including askew transformations (inadvertent alters to the genome), persevering altering impacts, genetic instruments in early stage and fetal turn of events, and longer-term wellbeing what’s more, security results.
Modifying one gene could have unanticipated and far and wide consequences for different pieces of the genome, which would then be passed down to people in the future.
Many believe genome changes to be exploitative, upholding that nature ought to be left to run its own course. Scarcely any contend that subsequent to allowing human germline Genome Editing under any circumstance would almost certainly prompt its obliviousness of as far as possible, to the rise of market-based selective breeding that would compound previously existing segregation, imbalance, and struggle.
It will end up being an instrument for choosing wanted attributes like knowledge and engaging quality.
It can likewise be utilized to dispense with risky types of bugs and not many trials are being conveyed out on mosquitoes however taking out an animal types, even one that doesn’t seem to have much biological esteem, could agitate the cautious equilibrium of environments.
That could have unfortunate results, such as upsetting the food web or expanding the danger that sicknesses like jungle fever could be spread by various species totally.
Current logical progressions show that CRISPR isn’t just an amazingly flexible innovation, it’s demonstrating to be precise and progressively protected to utilize.
In any case, a great deal of progress actually must be made; we are simply starting to see the maximum capacity of genome-altering apparatuses like CRISPR-Cas9.
Mechanical and moral obstacles actually stand among us and a future in which we feed the planet with designed food, dispose of genetic issues, or resurrect wiped out creature species.
CRISPR methodological enhancements incorporate treating cells with little molecules during altering to prod DSB fix away from NHEJ and toward HDR.
Controllable frameworks switch on Cas9 utilizing light or little particles, restricting its movement to decrease askew impacts.
Specialists are scouring the microbial world for new Cas-type compounds and altogether new genome-altering frameworks. “We’re actually distinguishing new particles with altering limit and we don’t completely comprehend the altering instruments we have,” Hennebold says. “We actually have a long way to go.”
The act of utilizing CRISPR to address illness causing changes is developing: Editas Medicine and Allergan declared human in vivo CRISPR-treatment preliminaries for an acquired visual impairment.
A likely obstacle to restorative CRISPR is the chance of human resistant reactions to its bacterial segments.
For example, a larger part of tried blood tests showed existing invulnerable reactions to Cas9, which is regularly taken from Staphylococcus or Streptococcus microorganisms.
Starting at 2012 productive genome altering had been created for a wide scope of test frameworks going from plants to creatures, regularly past clinical premium, and was turning into a standard trial system in research labs.
The new age of rodent, zebrafish, maize and tobacco ZFN-intervened freaks and the upgrades in TALEN-based methodologies vouch for the meaning of the strategies, and the rundown is growing quickly.
Genome altering with designed nucleases will probably add to numerous fields of life sciences from examining quality capacities in plants and creatures to quality treatment in people.
For example, the field of manufactured science which expects to design cells and creatures to perform novel capacities, is probably going to profit with the capacity of designed nuclease to add or eliminate genomic components and subsequently make complex frameworks.
Likewise, quality capacities can be contemplated utilizing immature microorganisms with designed nucleases.
Recorded underneath are some particular assignments this strategy can do:
• Designated quality change in Gene
• Gene treatment
• Making chromosome adjustment
• Study quality capacity with undifferentiated organisms
• Transgenic creatures
• Endogenous quality marking
• Designated transgene expansion
Designated quality change in creatures: The blend of ongoing disclosures in hereditary designing, especially quality altering and the most recent improvement in ox-like multiplication innovations (for example in vitro undeveloped organism culture) takes into consideration genome altering straightforwardly in prepared oocytes utilizing engineered exceptionally explicit endonucleases.
RNA-directed endonucleases: clustered routinely interspaced short palindromic rehashes related Cas9 (CRISPR/Cas9) are another instrument, further expanding the scope of strategies accessible.
Specifically, CRISPR/Cas9 designed endonucleases permits the utilization of various aide RNAs for synchronous Knockouts (KO) in one stage by cytoplasmic direct infusion (CDI) on mammalian zygotes.
Genome Editing Citations
- Genome Editing: Past, Present, and Future. Yale J Biol Med . 2017 Dec 19;90(4):653-659.
- The CRISPR tool kit for genome editing and beyond. Nat Commun . 2018 May 15;9(1):1911.
- A glance at genome editing with CRISPR-Cas9 technology. Curr Genet . 2020 Jun;66(3):447-462.
- CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu Rev Plant Biol . 2019 Apr 29;70:667-697.
- The promise and challenge of therapeutic genome editing. Nature . 2020 Feb;578(7794):229-236.
Share
Similar Post:
-

Non-coding DNA: Sequence, Definition, and Examples
Continue ReadingWhat is Non-Coding DNA?
The non-coding DNA portions are sections of DNA that don’t contain a quality and don’t code for a protein. These locales some of the time alluded to as “Junk DNA” are scattered all through the genome.
The noncoding locales get interpreted however are neither deciphered nor straightforwardly associated with the interaction of interpretation and henceforth no practical protein is created.
Be that as it may, a portion of these locales are thought to have known natural capacity. Certain locales additionally produce records that are included straightforwardly in RNA handling and interpretation, as opposed to being communicated into courier RNAs that encode proteins.
Its incorporates move RNAs (tRNA), ribosomal RNAs (rRNA), little atomic RNAs (snRNA), little nucleolar RNAs (snoRNA), and so on.
The number and variety of these non-coding districts remain basically obscure, even after the finishing of numerous genome arrangements.
A few inquiries like the quantity of non-coding qualities in a genome, their significance, their capacity inside a cell and regardless of whether these huge arrangements of qualities have gone undetected due to their failure to be converted into proteins have consistently stayed a secret.
"Non-coding DNA some of the time alluded to as "Junk DNA" are scattered all through the genome"
To resolve such inquiries, improvement of new precise quality disclosure approaches explicitly focused on non-coding areas is of most extreme need.
The possibility that these classes of qualities have stayed undetected is invigorating, if not distrustful. Non-coding areas are practically missing in bacterial genomes however makes up as much as 90% or a greater amount of the genome in higher organic entities.
Modern comparative genomics studies suggest that genetic variations between any two allied species are more because of the modifications in the non-coding regions slightly than in the protein-coding genes.
Transcription – the process which results in the formation of RNA molecules is the primary way in which genetic information affects a cell’s function.
Studies that employ metabolic labeling of newly synthesized RNA indicated that a vast proportion of nuclear DNA was actually transcribed, and the bulk of this heterogeneous RNA (hnRNA) never access the cytoplasm and hence did not get decoded into proteins.
Novel genomics and RNomic research have cross verified these findings.
Types of Non-coding DNA
I. Non-coding DNA Cis-and Trans-regulatory Components
Cis-regulatory components are arrangements that control the transcription of a close by gene. Numerous such components are engaged with the advancement and control of improvement.
Cis components might be situated in 5′ or 3′ untranslated districts or inside introns. Trans-regulatory components control the transcription of a far-off gene. Advertisers work with the transcription of a specific gene and are commonly upstream of the coding area.
Enhancer groupings may likewise apply exceptionally far off consequences for the transcription levels of genes.
II. Non-coding DNA: Introns
Introns are non-coding segments of a gene, transcribed into the forerunner mRNA succession, in any case eliminated by RNA grafting during the preparing to develop courier RNA.
Numerous introns give off an impression of being versatile genetic elements.
Investigations of gathering I introns from Tetrahymena protozoans demonstrate that a few introns seem, by all accounts, to be narrow minded genetic components, unbiased to the host since they eliminate themselves from flanking exons during RNA preparing and don’t create an articulation inclination between alleles with and without the intron.
Some introns seem to have critical natural capacity, conceivably through ribozyme usefulness that may direct tRNA and rRNA movement just as protein-coding gene articulation, obvious in has that have gotten reliant upon such introns throughout significant stretches of time;
For instance, the trnL-intron is found in all green plants and seems to have been upward acquired for a few billions of years, including in excess of a billion years inside chloroplasts and an extra 2–3 billion years earlier in the cyanobacterial progenitors of chloroplasts.
III. Non-coding DNA: Pseudogenes
Pseudogenes are DNA arrangements, identified with known genes, that have lost their protein-coding capacity or are generally presently not communicated in the cell.
Pseudogenes emerge from retrotransposition or genomic duplication of practical genes and become “genomic fossils” that are nonfunctional because of transformations that forestall the transcription of the gene, for example, inside the gene advertiser area, or lethally adjust the translation of the gene, like untimely stop codons or frameshifts.
Pseudogenes coming about because of the retro transposition of a RNA middle of the road are known as prepared pseudogenes; pseudogenes that emerge from the genomic stays of copied genes or deposits of inactivated genes are non-processed pseudogenes.
Transpositions of once useful mitochondrial genes from the cytoplasm to the core, otherwise called NUMTs, likewise qualify as one sort of normal pseudogene. Numts happen in numerous eukaryotic taxa.
While Dollo’s Law proposes that the deficiency of capacity in pseudogenes is conceivable lasting, quieted genes may really hold work for a few million years and can be “reactivated” into protein-coding sequences and a generous number of pseudogenes are effectively transcribed.
Because pseudogenes are attempted to change without developmental imperative, they can fill in as a valuable model of the sort and frequencies of different unconstrained genetic mutations.
To do again -repeat, transposons, and viral components: Transposons and retrotransposons are versatile genetic components.
Retrotransposon rehashed arrangements, which incorporate since a long time ago scattered atomic components (LINEs) and short mixed atomic components (SINEs), represent an enormous extent of the genomic groupings in numerous species.
Alu successions, named a short sprinkled atomic component, are the most plentiful versatile components in the human genome.
A few models have been found of SINEs applying transcriptional control of some protein-encoding genes.
Endogenous retrovirus groupings are the result of converse transcription of retrovirus genomes into the genomes of germ cells.
Transformation inside these retro-transcribed arrangements can inactivate the viral genome.
More than 8% of the human genome is comprised of (for the most part rotted) endogenous retrovirus groupings, as a component of the more than 42% portion that is conspicuously determined of retrotransposons, while another 3% can be distinguished to be the remaining parts of DNA transposons.
A significant part of the excess portion of the genome that is as of now without a disclosed beginning is required to have discovered its starting point in transposable components that were dynamic such a long time ago (> 200 million years) that arbitrary changes have delivered them unrecognizable.
Genome size variety in somewhere around two sorts of plants is for the most part the aftereffect of retrotransposon sequences.
IV. Non-coding DNA: Telomeres
Telomeres are locales of tedious DNA toward the finish of a chromosome, which give insurance from chromosomal weakening during DNA replication.
Late examinations have shown that telomeres capacity to help in its own dependability.
Telomeric rehash containing RNA (TERRA) are transcripts gotten from telomeres.
TERRA has been displayed to keep up with telomerase movement and stretch the closures of chromosomes.
Non-coding DNA Citations
- Functional variation and evolution of non-coding DNA. Curr Opin Genet Dev . 2006 Dec;16(6):559-64.
- A new method for species identification via protein-coding and non-coding DNA barcodes by combining machine learning with bioinformatic methods. PLoS One . 2012;7(2):e30986.
- Detecting the borders between coding and non-coding DNA regions in prokaryotes based on recursive segmentation and nucleotide doublets statistics. BMC Genomics . 2012;13 Suppl 8(Suppl 8):S19.
- The Genomic Code: A Pervasive Encoding/Molding of Chromatin Structures and a Solution of the “Non-Coding DNA” Mystery. Bioessays . 2019 Dec;41(12):e1900106.
- The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol . 2004;5(4):105.
- Rate variation in the evolution of non-coding DNA associated with social evolution in bees. Philos Trans R Soc Lond B Biol Sci . 2019 Jul 22;374(1777):20180247.
- Genome defense against exogenous nucleic acids in eukaryotes by non-coding DNA occurs through CRISPR-like mechanisms in the cytosol and the bodyguard protection in the nucleus. Mutat Res Rev Mutat Res . Jan-Mar 2016;767:31-41.
- Searching for functional genetic variants in non-coding DNA. Clin Exp Pharmacol Physiol . 2008 Apr;35(4):372-5.
- Genetic variation: Linear INSIGHTs into non-coding DNA. Nat Rev Genet . 2017 May;18(5):270-271.
- Protection of the genome and central protein-coding sequences by non-coding DNA against DNA damage from radiation. Mutat Res Rev Mutat Res . Apr-Jun 2015;764:108-17.
- The protective function of non-coding DNA in DNA damage accumulation with age and its roles in age-related diseases. Biogerontology . 2019 Dec;20(6):741-761.
Share
Similar Post:
-
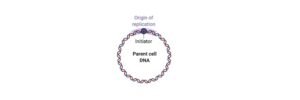
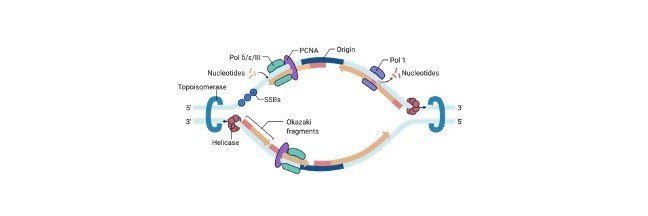
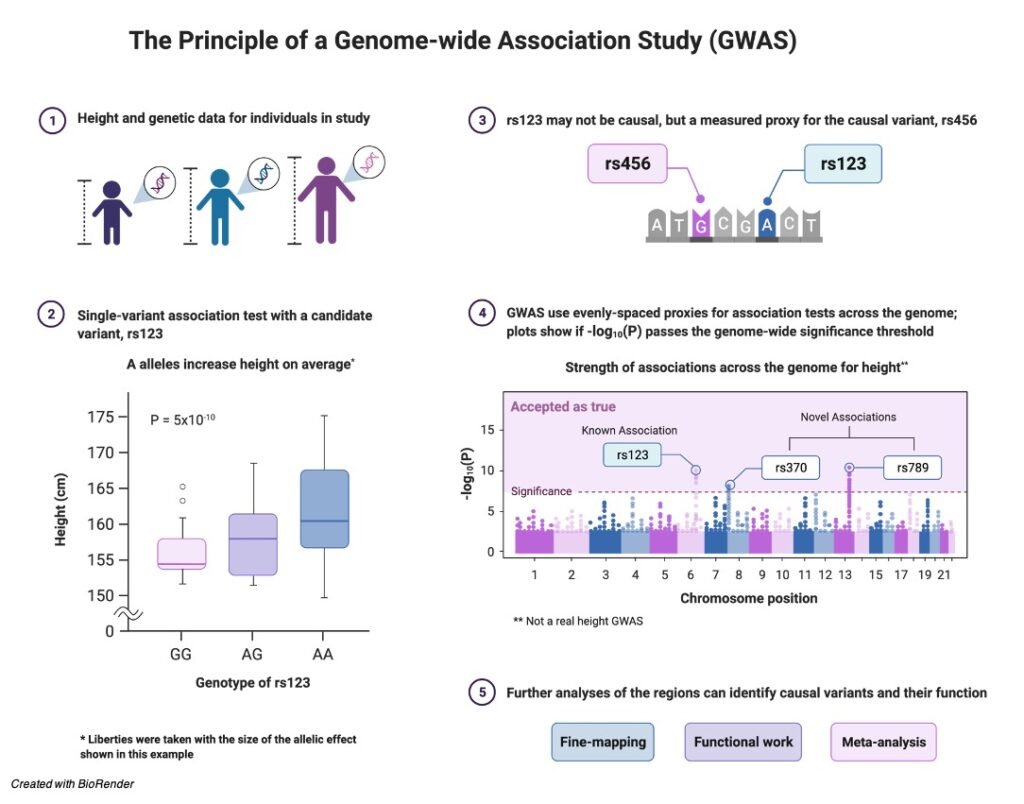
Origin of Replication: Definition, Structure, Diagram, and...
Continue ReadingOrigin of Replication
The origin of replication is nothing but a sequence of DNA, at which the replication is initiated on a chromosome, plasmid or a virus. For the small DNAs, which includes bacterial plasmids and other small viruses, where a single origin is sufficient.
Where as the organisms containing larger DNAs have many origins for replicating. The DNA replication is initiated in all of these.
In other cases, if the replication needs to be proceeded from a single origin, it takes a long time to replicate the entire mass of the DNA.
It is said that during each of the process of cell division, usually 30,000 to 50,000 activated origins of replication of DNA are present in humans and it does not remain clear till how these sites are selected and these sites are recognized by the replication factors.
Replication of DNA in the multicellular organisms should accommodate the variations in growth conditions and damage in the DNA.
It also should adapt the changes that is being occurring in the organization of the chromatin which is being associated with the process of cell differentiation and in development of the cell.
However, the selection of origin of replication in the metazoans was being occurred with variety of choices in identifying the cell and also in checking the conditions of the growth.
This also suggests that during the evolution processes, the replication uses its origin to become more controlled by the mechanism of epigenetic factors and mechanisms which affects the chromosomal dynamics and their expression than that of the DNA synthesis.
What is Origin of Replication?
The origin of replication, which is also known as replication origin is a particular sequence in a genome where the process of replication is initiated.
Propagation of a genetic molecules between the generations requires accurate and a timely duplication in the DNA by the process of semiconservative replication which occurs prior to the cell division to ensure that each daughter cells receives the full complement of the chromosomes.
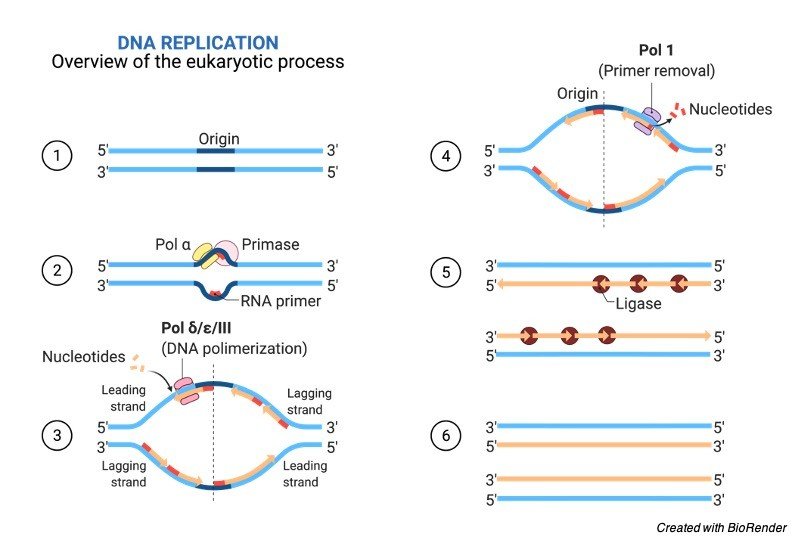
This replication process occurs both in prokaryotes and eukaryotes in their genetic material either DNA or RNA, where viruses contain mostly RNA; such as double stranded RNA viruses.
Production of double strands which have been initiated at a discrete site, which are termed as replication origin which further proceeds in a bidirectional manner till all the genomic DNA replicates.
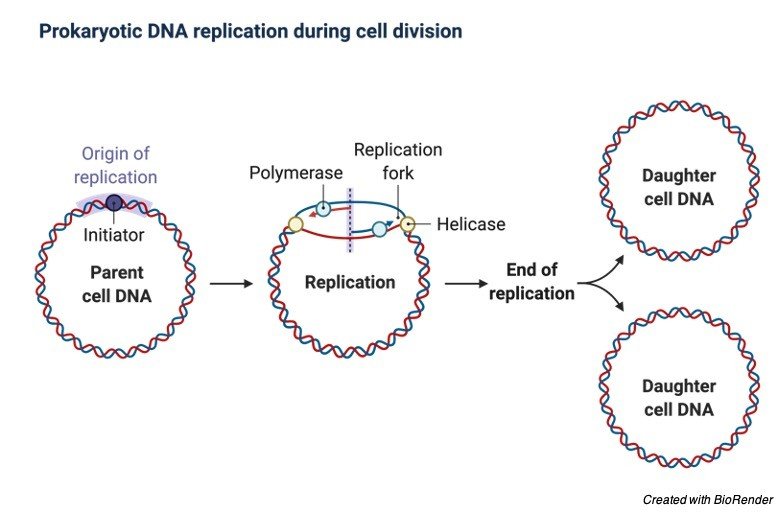
In spites the fundamental nature of this activities, organisms has evolved as divergent variants which control the onset of replication.
Though the replication process is specific in structure and in organizing the recognition sites varies from species to species, where only some common characters are shared.
Characteristics of Origin of Replication
An important need for the replication of DNA is it should occur in extremely high fidelity and should have exact efficiency in one cell per cell cycle which prevents the accumulation of the genetic alterations along potentially deleterious consequences for the survival of cell and in organismal viability.
Incomplete, erroneous or irregular replication of DNA gives rise to the mutations in the chromosomal polyploidy or aneuploidy of the chromosomes along with the variations in the copy number of the gene.
These leads to many diseases and disorders in the cell which also leads to tumor and cancer formations.
In order to ensure complete and accurate duplication in the genome the flow should be maintained correctly to inherit the accurate information to the newly produced cells.
All of these replications should be regulated in an accurate manner along with the cell cycle but it is also important for co ordinating the transcription and DNA repair.
In addition to this, it also originally sequences the commonly having high AT-content across all the cells , as it has number of repeats of adenine and thymine it is easy to separate their base stacking interactions which are not strong as those of guanine and cytosine.
Mechanism of Origin of Replication
DNA replication is divided into different stages. During the initiation the replication machineries which are commonly termed as replisomes are let to assemble on the DNA in a bidirectional fashion.
This assembling of loci constitutes the initiation sites for the process of DNA replication which is also termed as replication origins or origin of replication.
In elongation phases, these replisomes travel in the opposite directions with the replication forks, which unwinds the DNA helix and produces complementary daughter strands of DNA with the help of two parental strands which act as templates, Once the replication is completed, specific termination leads to the disassembly of the replisomes.
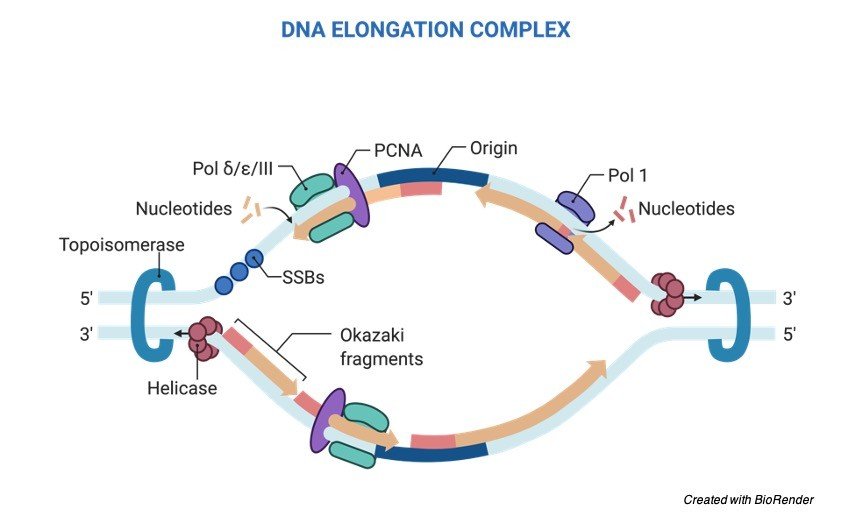
As longer the time takes for the duplication of the entire genome before the process of replication, we might assume that the particular location of sites of replication; where the replication starts does not taken into concern.
Although it has been shown that many of the organisms use the genomes regions which they have preferred for the process of replication as their initiation sites.
It is important to regulate the origin of replication because this step likely arises as there is a need to co ordinate the process of replication with the other processes which act on the shared template on the chromatin so that it helps to avoid the DNA strand breaks and the damage that occurs in the DNA.
Origin of Replication Citations
- Mapping the Single Origin of Replication in the Naegleria gruberi Extrachromosomal DNA Element. Protist . 2019 Apr;170(2):141-152.
- Chromosomal origin of replication coordinates logically distinct types of bacterial genetic regulation. NPJ Syst Biol Appl . 2020 Feb 17;6(1):5.
- Do Archaea Need an Origin of Replication? Trends Microbiol . 2018 Mar;26(3):172-174.
- Origin of DNA replication. PLoS Genet . 2019 Sep 12;15(9):e1008320.
Share
Similar Post: