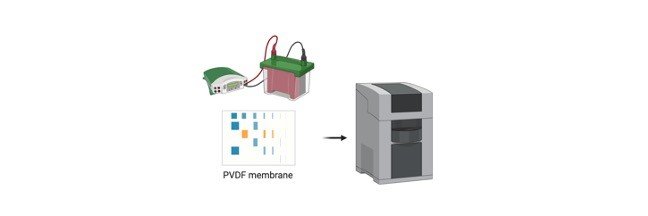
About Comet Assay
Several man-made chemicals find their way into the environment and pose health risk to human population.
These chemicals have been found to interact with the vital tissue macromolecules regulating the cellular functions leading to long lasting health disorders.
Acute and chronic exposure to environmental chemicals such as pesticide, metals, polycyclic aromatic hydrocarbons (PAHs), radiation and others have been shown to produce marked toxicity at the target sites.
Some of these chemicals affect the DNA, which is the carrier of inherited information and any gross change in its structure potentiates serious biological changes.
Hence there is a need to test the chemicals for their genotoxic potential before being released into the environment.
The conventional methods for evaluating genetic damage include chromosomal aberration, micronucleus assay, sister chromatid exchanges.
However, these are time consuming, resource intensive and require proliferating cell population.
Hence, newer and more sensitive test systems have now been introduced for assessing the genotoxicity of chemicals.
Fundamental Of Comet Assay
The comet assay and microgel electrophoresis (MGE) were first introduced by Ostling and Johanson in 1984.
This was a neutral assay in which the lysis and electrophoresis were done under neutral conditions.
The image obtained looks like a “comet” with a distinct head, comprising of intact DNA and a tail, consisting of damaged or broken pieces of DNA hence the name “Comet” Assay was given.
The extent of DNA liberated from the head of the comet was the function of the dose of damage.
The above neutral assay was modified by two groups, Singh and co-workers (1988) and Olive et al (1989). Singh et al used microgels, for electrophoresis under highly alkaline conditions (pH>13).
This enables the DNA super coils to get relaxed and unwind, which are then pulled out during application of electric-current which made possible detection of single strand breaks in DNA and alkali labile sites expressed as frank single strand breaks in individual cells.
This method was developed to measure low levels of strand breaks with high sensitivity.
Olive and co-workers conducted the electrophoresis under neutral or mild alkaline condition (pH 12.3) to detect single stranded breaks.
This method is optimized to detect a subpopulation of cells with varying sensitivity to drug or radiation.
Importance of Comet Assay
The single cell gel electrophoresis or comet assay is one such state-of-the-art technique for the measurement of DNA damage and repair in vitro in any eukaryotic cell and some prokaryotic cells.
This technique is rapid, non-invasive, sensitive, visual and inexpensive as compared to the conventional techniques and is a powerful tool to study factors modifying mutagenicity and carcinogenicity.
In addition, it combines the simplicity of biochemical techniques for detecting DNA single strand breaks (strand breaks and incomplete excision repair sites), alkali-labile sites and cross-linking with the single cell approach typical of cytogenetic assays.
Comet assay measures, double strand breaks (DBSs), single strand breaks (SSBs), alkali labile sites, oxidative DNA base damage, DNA-DNA/ DNA-protein/ DNA-Drug cross linking and DNA repair.
Comet Assay Principle
The assay works upon the principle that strand breakage of the super coiled duplex DNA leads to the reduction of the size of the large molecule and these strands can be stretched out by electrophoresis.
Comets form as the broken ends of the negatively charged DNA molecule become free to migrate in the electric field towards the anode.
Two principles in the formation of the comet are:
1. DNA migration is a function of both size and the number of broken ends of the DNA
2. Tail length increases with damage initially and then reaches a maximum
Advantage of Comet Assay
1. It is a non-invasive technique.
2. It requires <10,000 cells and collection of data at the level of the individual cell, allowing for more robust types of statistical analyses.
3. Counting of 50-100 cells per individual/ treatment group through computerized image analysis software gives a complete analysis.
4. Any eukaryotic cell population (nasal and buccal mucosal cells, epithelial cells, male germ cells, fine needle biopsy) is amenable to analysis.
5. Duration of this experiment is few hours only whereas conventional cytogenetic assays require few days.
6. Single strand breaks (SSBs) and alkali labile lesions (capable of being transformed into SSBs under alkaline conditions) in the DNA of individual cells can be assessed.
Materials Required for Comet Assay
Frosted slides
Coverslips
Lymphocyte Separation Medium
Hydrogen peroxide (H2O2): 25 μM and 50 μM
Phosphate Buffer Saline (PBS) (i.e. 137mM NaCl, 2.7mM KCl, 10mM Na2HPO4.2H2O and 2 mM KH2PO4, pH 7.4).
Dulbecco’s modified Eagle Medium (DMEM)
Fetal Bovine Serum (FBS)
0.4% Trypan blue dye
Normal melting point agarose (NMPA)
Low melting point agarose (LMPA)
Propidium iodide
Lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Trizma base).
Neutralising Buffer (Add 0.4 M Tris to ~800 ml distilled water. Adjust the pH to 7.5 using conc. HCl to 1000 ml with distilled water).
Electrophoresis buffer (Add 30 ml 10N NaOH and 5 ml 200 mM EDTA to 1000 ml with distilled water, mix well. Ensure pH > 13 before use).
IX-51 inverted research microscope
Horizontal gel electrophoresis units
Sorvall Centrifuge
Comet Assay Procedure
Isolation of lymphocytes:
Blood was collected in microfuge tubes containing heparin (10 IU heparin per ml of blood)
Blood was diluted with 1X PBS in the ratio of 1:3 (Blood: PBS) and layered on lymphocyte separation medium (LSM) (1 ml of LSM per 3 ml of diluted blood)
Tubes were centrifuged at 400 g for 15 minutes at 15oC.
Lymphocyte layer was collected and diluted with 1X PBS in the ratio of 1:1.
Again it was centrifuged at 400 g for 10 mins at 15oC to get the lymphocyte pellet.
Pellet was resuspended in 1X PBS.
Estimation of cell viability using trypan blue exclusion assay. The standard trypan blue assay was carried out on isolated lymphocytes as mentioned in earlier protocol for cell counting.
Chemical treatment of cells:
An aliquot of cells were treated with two different concentrations of H2O2 (25 μM and 50 μM) for 15 minutes and the other aliquot of untreated cells were used as negative control.
A layer of 1% normal melting point agarose (NMPA) was prepared on frosted-end slides and slides were kept overnight at 37oC.
After chemical treatment, cells (1 x 106 cells/ml) were mixed with 1ml of 2% low melting point agarose (LMPA).
The suspension of 200 μl LMPA and cells was layered on to the precoated slides and covered with cover slips.
The layer was allowed to solidify at 4oC for 15 mins.
After solidification of the layer, cover slips were removed.
Slides were immersed in cold lysis solution (pH 10) and kept at 4C for 3 hours.
Slides were washed gently with alkaline electrophoresis buffer (pH 13) for 15 mins, twice.
This helps in allowing denaturation of DNA.
Subsequently, slides were transferred to an electrophoresis tank with fresh alkaline electrophoresis buffer and electrophoresis was performed at field strength of 20 V/ 250 mA for 25 min at 4C.
Slides were neutralized in chilled distilled water for 5 min and stained with 5 μg/ml propidium iodide for 20 mins.
DNA damage Analysis in Comet Assay
For visualization of DNA damage, observations are made with PI stained DNA using a 10X/20X objective on a fluorescent microscope.
Any image analysis system may be suitable for the quantification of SCGE data.
CASP, freely downloadable software, is used to assess the quantitative extent of DNA damage in the cells by measuring the length of DNA migration and the percentage of migrated DNA.
The software calculates the tails moment and olive tail moment.
Generally, 50 to 100 randomly selected cells are analyzed per sample.
The softwares are designed to differentiate comet head from tail and to measure a variety of parameters including tail length; % of total fluorescence in head and tail; and ‘tail moment’, calculated in different ways but essentially representing the product of tail length and relative tail intensity.
Percent DNA in tail is linearly related to DNA break frequency up to about 80% in tail, and this defines the useful range of the assay.
Tail length tends to increase rapidly with dose at low levels of damage, but soon reaches its maximum.
It is therefore the most sensitive parameter at near-background levels of damage.
Tail moment is an attempt to combine the information of tail length and tail intensity, but suffers from lack of linearity.
DNA damage Analysis with CASP Software in Comet Assay
The CASP software can be downloaded from http://www.casplab.com
The software CASP generates the frame to limit the comet area, the line to limit the tail length, the circle to identify the comet head and the cross to identify the head centre.
The small rectangle at the bottom of each picture is generated by CASP as background reference.
Comet Assay Parameters
1. % Head DNA = (Head optical intensity/ (Head optical intensity + Tail optical intensity)) X 100
2. % Tail DNA = 100- % Head DNA.
3. Tail length = Tail length is the distance of DNA migration from the body of the nuclear core and it is used to evaluate the extent of DNA damage.
4. Tail length = Tail extent (Tail from centre) + Head extent / 2
5. Olive tail moment = Tail moment is defined as the product of the tail length and the fraction of total DNA in the tail.
Tail moment incorporates a measure of both the smallest detectable size of migrating DNA (reflected in the comet tail length) and the number of relaxed/ broken pieces (represented by the intensity of DNA in the tail).
6. Olive Tail Moment = (mean Tail- mean Head) X % Tail DNA/ 100 7. Extent Tail Moment = Tail length X % Tail DNA / 100
Comet Assay Citations:
Share