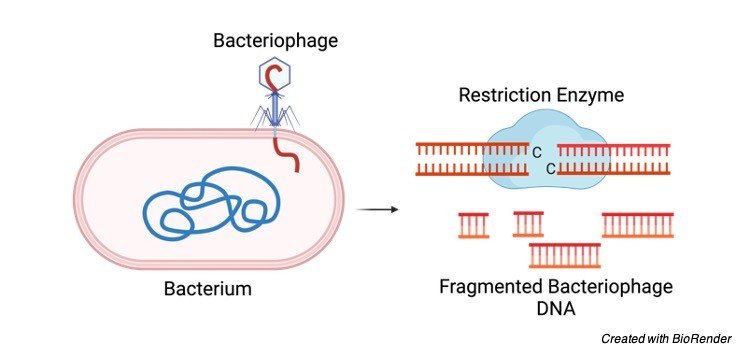
What are Restriction Enzymes?
Restriction enzymes also known as restriction endonuclease is a protein synthesize by bacteria is responsible for the cleavage of DNA at a specific site.
The site of cleavage is known as restriction site.
They are also known as ‘molecular scissors’ due to their ability to identify specific base sequence on DNA and then incising them at a particular location.
This enzyme plays a pivotal role in defending the live bacteria from bacteriophages by identifying and severing the DNA of bacteriophage at the restriction site. Thus, killing the phage.
In the modern science, restriction enzyme plays crucial role in the field of genetics, where it is used as powerful tool to cut the DNA or RNA at particular site.
Moreover, it is uncomplicated to extract it from the bacteria and to use it in the laboratory.
They identify tiny and discrete nucleotide sequences in the DNA which is commonly known as recognition sequences.
On recognition of the DNA sequence, the enzyme hydrolyzes the bond between two nucleotides and cleave the DNA molecule.
However, the bacteria which carries this enzyme prevents self-degradation by incorporation of the methyl group at the adenine or cytosine bases within the recognition sequence with the assistance of enzyme methylases.
Source of Restriction Enzymes
Naturally these can be extracted from the bacterial cells.
Each bacteria generate their own unique restriction enzymes while each enzyme identifies only particular type of restriction sequence.
Also, they recognize remote DNA and severe them at particular location along the molecule.
Recognition Sites of Restriction Enzymes
Any particular location where the DNA double helix is severed into fragments is known as restriction sites.
This DNA sequence identified by endonuclease is called palindromes.
Palindromes can be defined as the base sequences that read in similar manner on both the strands but in opposite directions.
For instance, if the sequence of DNA on one stand reads GAATTC from 5’ → 3’direction, the sequence on the opposite strand has to read CTTAAG in the 3’ → 5’ direction.

However, when both the strands scrutinize in the 5’ → 3’ direction, the sequence remains the same.
Thus, the palindromic sequence appears as
5’ GAATTC 3’
3’ CTTAAG 5’
Moreover, there exist point of symmetry within the palindrome. In the above example, AT/TA is the center between the sequence.
It was recognise that restriction enzymes severe the DNA molecule throughout this point of symmetry.
Sometimes, enzymes cut straight through the molecule at the symmetrical axis creating blunt ends.
However, the enzyme which cuts both the strands at a distant from the point of symmetry between the similar 2 bases are of more value resulting in staggering break of the DNA.
Types of Restriction Enzyme
Thus, according to the recognition site, three types of restriction enzymes are categorized as follows:
Type I Restriction Enzyme
They cut the DNA away from the recognition sequences. But, they are unable to create individual restriction fragments making them less valuable from the practical point of view.
They form complex, multi-subunit restriction and modification enzymes.
Upon genomic analysis, it was observed that they are common and has significant biochemical interest, which at the beginning considered rare.
Type II Restriction Enzyme
They cut at particular position within the range of the restriction sites.
It was observed that, the fragments formed are separate and exhibit gel banding pattern.
These enzymes show pivotal role in the laboratories wherein they are exclusively utilized for DNA analysis and gene cloning.
These are considered as family of unrelated proteins.
They are extracted from bacteria and accordingly termed after that particular bacterial species.
For example, enzyme isolated from the E.coli is called as EcoR1.
These enzymes creates 2 types of cuts to the DNA:
• Blunt ends: It is seen when the enzyme cut through the DNA from centre of the recognition sequence
• Sticky ends: when it creates an overhang of the molecules.
Type III Restriction Enzyme
The protein forming this type has multiple function which have 2 subunits with types – Res and Mod.
It is a modification methyltransferase.
The specific DNA sequence required for the system is identified by Mod subunit.
Mechanism of Restriction Enzymes
On recognizing the particular sequence, it incises the DNA molecule by hydrolysis of the bond between the adjacent nucleotides wherein water is consumed to split the bond chemically.
Since the DNA is double stranded, to snip it, the restriction enzyme severe it twice through each sugar-phosphate backbone.
Applications of Restriction Enzymes
A. Restriction Fragment Length Polymorphism (RFLP) Technique
In molecular biology, this technique is used to specifically locate the site of genes within a DNA sequence by severing the DNA into minute fragments.
Thereafter these fragments are studied to distinguish individuals, populations and species.
B. Gene Cloning
In the cloning procedure, a gene sequence is cut using the restriction enzyme which is then inserted into a plasmid.
The restriction enzyme also severe the plasmid resulting in creation of single-stranded overhangs or sticky ends of the DNA over plasmid.
The two loose DNA molecules are ligated back using another protein called as DNA ligase, thus, forming a single DNA molecule.
C. Gene Mapping
Restriction endonucleases are used to modify the DNA.
Moreover, identifying a specific site and severing them at that particular site, make them an indispensable instrument to create a physical map of DNA, thus mapping the whole human genome.
With the use of each fragment, further analysis and sequencing can be done.
D. Gel Electrophoresis
The added advantage of the restriction enzyme is that it identifies the same distinct base sequence regardless of the source of the DNA.
Thus, it creates a stable landmark throughout the regular DNA sequence.
This will lead to splitting of the DNA molecule into particular specks where it can be separated with help of gel electrophoresis.
Significance of Restriction Enzymes
With the inception of technology into the biological world, we are directed towards molecular biology where every biochemical process is observed with great detail.
In this era, restriction enzymes are an indispensable device in the genetic industry.
Till date, there were over 19,000 enzymes have been identified and still more will come in the future.
Each one of them have specific target location in a DNA sequence thus, making them more specific.
In the commercial world, there are 4 types of restriction enzymes available, out of which type II restriction enzymes are widely accepted in recombinant DNA technology.
In the present situation, more than 300 type II restriction enzymes are available and very less number of other types are characterised.
However, after studying these enzymes, valuable information has been shared regarding the DNA-protein interactions and catalysis, plasticity of protein domain, protein family relationships and control of restriction activity.
ribosomal RNA (rRNA): Definition, Classification, and Function
Restriction Enzyme Citations
- Restriction enzyme-mediated DNA family shuffling. Methods Mol Biol . 2014;1179:175-87.
- Restriction enzyme mining for SNPs in genomes. Anticancer Res . Jul-Aug 2008;28(4A):2001-7.
- The other face of restriction: modification-dependent enzymes. Nucleic Acids Res . 2014 Jan;42(1):56-69.
- Restriction endonuclease digestion of DNA. Methods Mol Biol . 1993;18:427-31.