What is DNA Replication?
DNA replication is the process by which a molecule of DNA is duplicated. When a cell divides, it must first duplicate its genome so that each daughter cell winds up with a complete set of chromosomes. DNA is made up of a double helix of two complementary strands.
The double helix describes the appearance of a double-stranded DNA which is thus composed of two linear strands that run opposite to each other and twist together to form. During replication, these strands are separated.
Each strand of the original DNA molecule then serves as a template for the production of its counterpart, a process referred to as semiconservative replication.
As a result of semi-conservative replication, the new helix will be composed of an original DNA strand as well as a newly synthesized strand. Cellular proofreading and error-checking mechanisms ensure near perfect fidelity for DNA replication.
What is Replication Fork?
DNA replication occurs during the synthesis (S) phase of the cell cycle and starts at predefined DNA sequences known as replication origins; this start is also known as origin firing.
Once the origins of replication have fired, the DNA replication proteins organize into a structure called the replication fork (RF), where a group of proteins coordinate DNA replication.
It is called a fork because the simplified structure resembles a two-tined fork, but its function is hardly simple, since it is the location of dynamic activity among a large group of proteins.
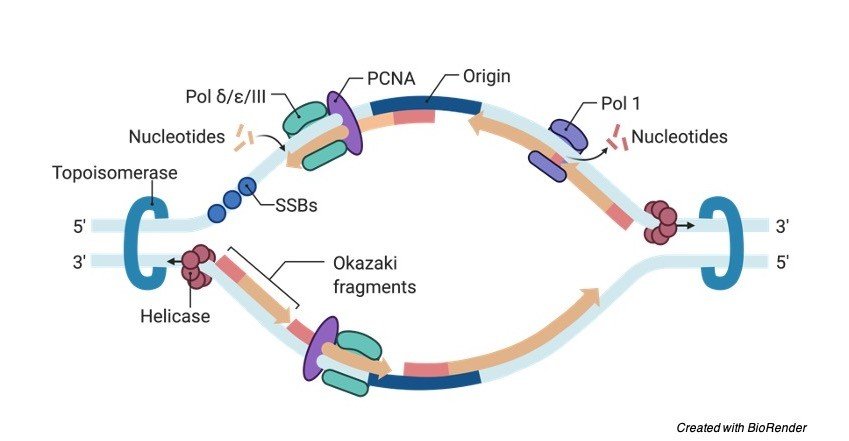
What Happens at the Replication Fork?
Two main activities happen at the fork: DNA unwinding and DNA synthesis. The RF unwinds the unreplicated DNA ahead of it through a helicase enzyme complex. As their name suggests, helicases modify the structure of the DNA helix and promote unwinding and separation of the two DNA strands.
The second activity of DNA synthesis at the RF is undertaken by DNA polymerase. This enzyme links together, or polymerizes, DNA bases in the correct sequence using the template DNA strand, and it generates two copies of the genome that are later divided into daughter cells in metaphase, or M phase.
What is Origin of DNA replication?
Replication always starts at specific locations on the DNA, which are called origins of replication and are recognized by their sequence. E. coli, like most bacteria, has a single origin of replication on its chromosome.
The origin has mostly A/T base pairs (which are held together by fewer hydrogen bonds than G/C base pairs), making the DNA strands easier to separate.
Specialized proteins recognize the origin, bind to this site, and open up the DNA. As the DNA opens, two Y-shaped structures called replication forks are formed, together making up what’s called a replication bubble. The replication forks will move in opposite directions as replication proceeds.
Mechanism of DNA Replication
We all known that DNA is a double stranded structure with two complementary strands. Where two linear strands run opposite to each other in a twisted manner.
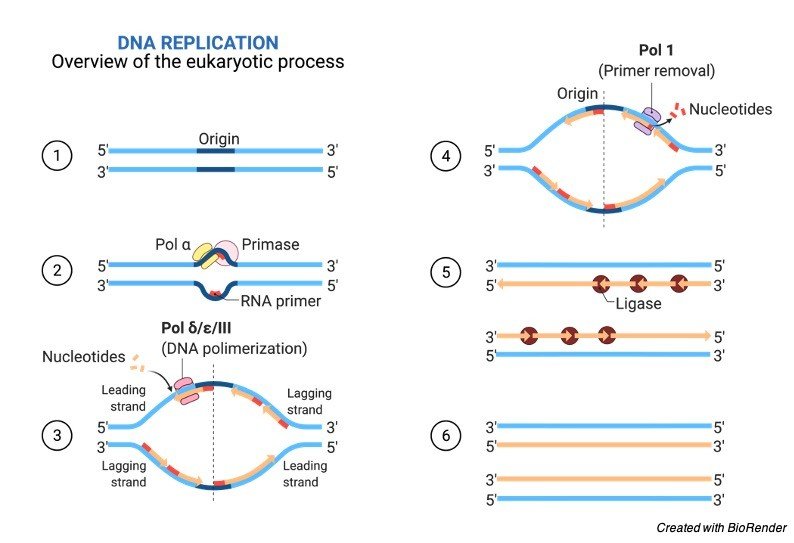
During the process of replication these two strands are separated and the separation initiated at the replication fork during the step of elongation.
Replication Fork
The replication fork is a structure which is formed during the process of DNA replication.
It is activated by helicases, which helps in breaking the hydrogen bonds, and holds the two strands of the helix.
The resulting structure has two branching’s which is known as prongs, where each one is made up of single strand of DNA.
These two strands serve as a template for the leading and lagging strands, which will be created as DNA polymerases which matches the complementary nucleotide with the templates, then the templates are as usual properly referred to as the leading strand template and the; lagging strand template.
DNA is being read by the DNA polymerase in the 3’ to 5’ direction which means that a new strand is being synthesised in the 5’ to 3’ direction.
Since the leading and the lagging strand templates which are oriented in the opposite directions at the replication fork, a major issue is in synthesis of the lagging strand whose direction of synthesis is opposite to the direction of the replication forks growth.
Leading Strand in Replication Fork
The leading strand is one of the strands where the new DNA is synthesised along the same direction of the growing replication fork. This method of DNA replication occurs in a continuous manner.
Lagging Strand in Replication Fork
The lagging strand is one other type of strand where the new DNA is produced in the direction opposite to that of the growth of replication fork.
Because of this type of orientation, lagging strand replication is considered more complicated on comparing with the leading strand.
In the same manner the DNA polymerase in this strand is seen to lag behind, on the other strand Whereas the lagging strand is produced in a short and separated segment.
In Lagging strands template an enzyme primase extends the primed segments, which forms the ookazagi fragments.
Following this the RNA primers are removed and then it is replaced with the DNA and their fragments which are then joined by the enzyme DNA ligase.
Dynamics at the Replication Fork
In all the cases, the enzyme helicase composes of six polypeptides which wraps around one of the strands of the DNA, that is being replicated.
The two polymerases are then bound to the enzyme helicase hexamer. In eukaryotes this helicase wraps around the leading strand and in prokaryotes it is winded around the lagging strand.
Due to this helicase unwraps the DNA, at the replication fork, The DNA is now forced to rotate and this process results in building up the twists in the DNA ahead.
This building up of forms a torsional resistance which results in eventually halting the process of replication of fork. Topoisomerases are the enzymes which temporarily breaks the strands of DNA, by relieving the tension which are caused by unwinding the double strands of DNA helix.
Topoisomerases which also includes DNA gyrase which is achieved by adding negative supercoils to the DNA helix.
Bare single stranded DNA has the capability to fold back on itself that tends to form a secondary structure., these structures further interfere with the movement involved by DNA polymerases.
So, in order to prevent this single strand binding proteins are bind to the DNA until the second strand is produced, which prevents the formation of secondary structures.
Double stranded DNA is usually coiled by the histones during replication which play an important role in regulating the gene expressions, so that the replicated DNA must be coiled around the histones at the same place as the original DNA.
In order to ensure this histone proteins, dis assembles the chromatin before it has been replicated and further it replaces the histones in an appropriate place. The steps involved in reassemble must be appropriate.