Author: Admin
-
Repetitive DNA Sequences: Definition, Types, and Meaning
Continue ReadingRepetitive DNA Sequences
The analysis of genetic diversity and relatedness within the species is surviving as a major theme of research for most of the researchers.
The availability of whole genome sequencing, which increases the number of species, focus have been shifted to the molecular development which is based on the DNA or protein polymorphism.
DNA sequences usually originate and undergoes an evolutionary metamorphosis; which is being used as a powerful tool for the genetic markers to characterise the genome among wide variety of species.
This type of analyses is known as DNA fingerprinting or profiling or genotyping.
These techniques were developed by Jeffreys et al in the year 1985, He also demonstrated the short repeat sequences tandemly arranged in the genome and he also identified that each organism has a unique pattern of arrangements.
DNA finger printing has then become the most powerful tool in solving the problems related to paternity such as crimes and genetic disorders etc.
In addition to all this, techniques of molecular biology which involves the isolation of genes tagged with minisatellites has became the most powerful weapon for analysis of genome.
The term repetitive sequences which are also known as DNA repeats of repetitive DNA is defined as the DNA fragments which are present as multiple copies within the genome.
These sequences exhibit a high degree of polymorphism as a result of variations in their number of repeats which causes mutations and thus affects several mechanisms in proper functioning of a body.
Features of Repetitive DNA Sequences
Repetitive DNA is generally defined as the sequence of DNA which are repeated in a genome.
These sequences do not code for the proteins. One of this class is termed as highly repetitive DNA.
DNA consists of short sequences having about 5-100 nucleotides, which are repeated more than thousand times in a single stretch; It also includes a satellite DNA which comes under the other class and consists of a longer sequence.
These longer sequences consist about 150 to 300 nucleotides which are dispersed throughout the genome evenly.
Characterstic of Repetitive DNA Sequences
The repetitive DNA is composed of the repeated sequences as such the name implies. It was originally identified by Cot-value which is derived from the kinetic studies of the renaturation of DNA.
These repeated sequences occur as a multiple copy of nucleic acids in the genomes. These repeats occurs both in DNA and in RNA.
Generally, in repetitive DNA, DNA repeats occurs during the stretches that occurs in the genome either as tandem or in the interspersed form along the genome.
These repeats can be found in various organisms including humans. In humans, there are over two-third of the repetitive DNA in the genome.
One of the class of repetitive DNA is termed as highly repetitive DNA which is found in short sequences consisting of about 5 to 100 nucleotides, which are found in a form of repeated sequence along a long stretch.
It is typically composed of 3 to 10% of the genomic DNA, among which satellite DNA is present predominantly.
Another class which is termed as moderately repeated DNA composes almost 25 to 40% of the genome.
This moderately repetitive sequence consists of about 150 to 300 nucleotide sequences that are dispersed throughout the length of the genome.
It also includes alu sequences and the transposons.
Types of Repetitive DNA
Repetitive DNA can be classified into two types as Highly repetitive sequences and moderately repeated sequences.
I. Highly Repetitive DNA
These are generally the short sequences of the genomes, which account for about10% of the genome containing 5 to 10 base pairs.
This type usually occurs as a tandem repeat. But they are interspersed among different non-repetitive sequences.
Almost all the sequences belonging to this class are located in the region of heterochromatin in the chromosomes.
Highly repetitive sequences interact with a specific protein that are involved in organising the chromosome while pairing at the stages of meiosis and recombination.
II. Satellite DNA
Satellite DNA are mostly represented by the monomeric sequences which are usually less than the 2000 base pairs.
They are tandemly reiterated up to one lakh copies per haploid organism.
These are located in the pericentromeric or in the telomeres in the heterochromatin regions.
Satellite DNA comprises of about 1 to 65% of the total DNA in most of the organism including plants and all animals and also in some of the prokaryotes.
III. Moderately Repetitive DNA Sequences
This includes a short sequence of about 150 to 300 base pairs and accounts for about 40% of the total genome.
These are found dispersed throughout the euchromatin which have 1000-100000 copies per haploid genome.
These sequences are especially involved in the regulation of gene expression.
The long sequences present in this is about 5 kilo base pairs.
This can be classified into two classes as tandem repeats and interspersed repetitive DNA.
IV. Tandem Repeats
Tandem repeats comprise of repeated arrays counting from two to several thousand sequences that are arranged in a head to tail fashion.
These are further divided into 3 categories depending on the length and copy number of the basic repeat units in the genomic localization.
V. Interspersed Repetitive DNA
These sequences are found scattered throughout the genome and they are raised by transposition, which have the capability to jump from one end to the other in the genome.
Although the individual units of this sequences are not clustered, it accounts for about 45% of the human genome.
These are also known as non-coding DNAs. During the mechanism of transposition, these interspersed are categorised into two classes.
1. RNA Transposons
These are also commonly called as retroelements, which were found in the eukaryotic genomes which requires reverse transcription for initiating their activity.
Based on their structural relationship RNA transposons are further classified into two sub classes as. LTR elements and Non-LTR elements.
2. DNA Transposons
DNA transposons usually do not require RNA intermediate and transpose in a direct DNA to DNA form.
Where as in eukaryotes, DNA transposons are found less commonly than the retrotransposons, as they have a special place in the family of plants as plant DNA transposons.
There are two types of DNA transposons which requires enzymes that are coded by the genes, within the transposons.
Repetitive DNA Sequences Citations
- The repetitive DNA content of eukaryotic genomes. Genome Dyn . 2012;7:1-28.
- Repetitive DNA loci and their modulation by the non-canonical nucleic acid structures R-loops and G-quadruplexes. Nucleus . 2017 Mar 4;8(2):162-181.
- Replication Through Repetitive DNA Elements and Their Role in Human Diseases. Adv Exp Med Biol . 2017;1042:549-581.
- Why repetitive DNA is essential to genome function. Biol Rev Camb Philos Soc . 2005 May;80(2):227-50.
Share
Similar Post:
-

What is DNA Polymorphism? Definition, Types, and...
Continue ReadingDNA Polymorphism
Human genome comprises of about 3 billion bases. There are about 10 million single nucleotide polymorphisms. The DNA sequences will carry genetic code as the information regulator for carrying out genetic information from parents to the offspring.
We can sort out the difference between animal or a plant or a Human as they are looking phenotypically different, which means that there are variations in their genomes in each of the organisms.
Any change in the sequence of the genetic material results in mutation or change in the genetic information, which in turn brings out a change in the phenotypic expression and also in the biological functions.
This change in sequence of a nucleotide or in genetic material is commonly known as mutation.
When we found a frequency of mutation occurring greater than 2 percentage in the population then is known as DNA polymorphism.
Hence DNA polymorphism can be defined as the DNA which exists in more than one form, having a frequency of greater than 2 percentage.
DNA Polymorphism Features
DNA polymorphism is referred to the different forms of DNA sequences within individual or among populations.
Polymorphism at a DNA level involves a wide range of variations from single base pair to many base pair alterations and also in repeated sequences.
DNA polymorphisms are considered as endless. These DNA polymorphisms are produced by changes in the sequence of the nucleotides or in the length of the base pairs. Or in length of the fragments.
Nucleotide DNA Polymorphisms
Variations in the genomes at a DNA level may be present in many forms including the single nucleotide polymorphisms, variable number of tandem repeats in mini and micro satellites, transposable elements, structural alterations and variations in the copy number. These variations occur either in a nucleus or in the mitochondria.
There are two major sources for these variations, one is mutations which results as a chance processes or the ones which have been induced by external agents like radiations or other environmental factors.
The second is the recombination which is formed by inheritance from parents followed by the offspring.
Generally, the genomes of the humans can be divided into different parts based on the functional elements as coding and the non-coding regions.
The coding regions mostly contains the DNA sequences which determines the primary amino acid sequences.
On the other hand, non-coding DNA consists of the sequences of the DNA which does not have any functions.
Such sequences exists either as a single copy or in multiple copies which are known as repetitive DNA.
These regions which does not code for proteins tend to have more polymorphisms.
Types of DNA Polymorphism
DNA polymorphisms can be classified into various types as follows;
1. Single Nucleotide DNA Polymorphisms
When a change takes place in a single base, it leads to High density natural sequence variations in the genome of an individual.
Single Nucleotide Polymorphisms mostly occur when there is an error such as deletion or insertion or substitutions of the bases of the nucleotides.
SNPs are considered as a prominent source in variation in the human genome which serves as an excellent genetic marker.
Whereas some regions of the genomes are richer in single nucleotide polymorphisms while others are in a normal condition.
SNPs are mostly present in the non-coding regions of the genomes and they have no direct impact on the phenotype of the particular, but their role remains active and might causes different consequences at the phenotypic level.
2. Insertion or Deletion Polymorphisms
This is one of the types of variation in a DNA; in which the specific nucleotide sequences of the various lengths ranging from one to more than 100 base pairs are deleted or inserted. These variations are spread widely across the genomes.
3. Polymorphic Repetitive Sequences
DNA repeats are often classified as interspersed repeats or tandem repeats, which comprises over two third of the human genome.
Whereas interspersed repeats are found dispersed across the genomes as intergenic and it includes pseudo genes and transposons.
On the other hand, Tandem repeats are found adjacent to each other and involves only a few copies of base pairs.
Tandem repeats are often found in Telomeres and centromeres. Tandem repeats are arranged in a head to tail manner depending upon the size of each repeat unit.
Whereas satellite repeats can be divided into microsatellites and minisatellites. Where, microsatellites have a repetitive sequence which is longer than hundred base pairs and they are also larger than the tandem repeats, located on one or many chromosomes.
Minisatellites are stretches of the DNA and they are characterised by normal length patterns which has ten to hundred base pairs, but usually less than 50 base pairs are only present.
Macrosatellites are also known as short tandem repeats, because the repat units are less than 10 base pairs.
4. Structural and Copy Number Variations
These are another type of frequent genome variability. Structural and copy number variations are denoted as CNVs, Which involves segments of DNA less than one kilo bases, copy number polymorphism is also known as duplication or deletion and it involves less than one kilo base of DNA.
Where as the intermediate sized structural variant and a structural variant involves more than eight to forty kilo bases.
This is usually referred as CNVs or balanced structural rearrangement, also known as inversion.
DNA Polymorphism Citations
- DNA polymorphisms in human population studies: a review. Ann Hum Biol . May-Jun 1987;14(3):203-17.
- Pathogenic mutations and non-pathogenic DNA polymorphisms in the most common neurodegenerative disorders. Folia Neuropathol . 2007;45(4):164-9.
- DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics . 2009 Jun 1;25(11):1451-2.
- A DNA polymorphism discovery resource for research on human genetic variation. Genome Res . 1998 Dec;8(12):1229-31.
- Mitochondrial DNA polymorphisms and extraversion. Am J Med Genet B Neuropsychiatr Genet . 2004 Jul 1;128B(1):76-9.
- Colorful DNA polymorphisms in humans. Semin Cell Dev Biol . Jun-Jul 2013;24(6-7):562-75.
- New application for DNA polymorphism. N Engl J Med . 1987 Feb 19;316(8):478-80.
- DNA polymorphism of schistosomes and their snail hosts. Mem Inst Oswaldo Cruz . Mar-Apr 1995;90(2):211-3.
- The application of capillary electrophoresis for DNA polymorphism analysis. Methods Mol Biol . 2001;162:3-26.
- DNA polymorphism and the study of disease associations. Hum Genet . 1988 Apr;78(4):299-312.
Share
Similar Post:
-

Gel Electrophoresis: Definition, Principle, and Application
Continue ReadingWhat is Gel Electrophoresis?
Before knowing about Gel electrophoresis, let us know few basics of what electrophoresis is?
Electrophoresis is a process used for the separation of macro and micro molecules in an electric field by applying charges at both the extents.
The mixture of substances is spread in the supporting film. The supporting films are placed in a salt solution filled in a container, where one container holds a cathode and the other carries an anode.
When the electric current is allowed to pass through this field, negatively charged molecules move towards the anode and positively charged molecules move towards the cathode.
Depending on the type of supporting films, they are classified as different types of electrophoresis. The supporting films may be of paper, agar, gel or starch.

Horizontal Gel electrophoresis
Depending on the type of substances to be separated the type of electrophoresis varies.
Moving boundary electrophoresis and Gel electrophoresis is used for the separation of proteins.
Discontinuous electrophoresis is used for the separation of proteins from plasma membrane.
SDS-PAGA (Sodium dodecyl Sulphate – Polyacrylamide gel electrophoresis) is used for the separation of macro molecules according to their size like membrane proteins and protein element of cytoskeleton.
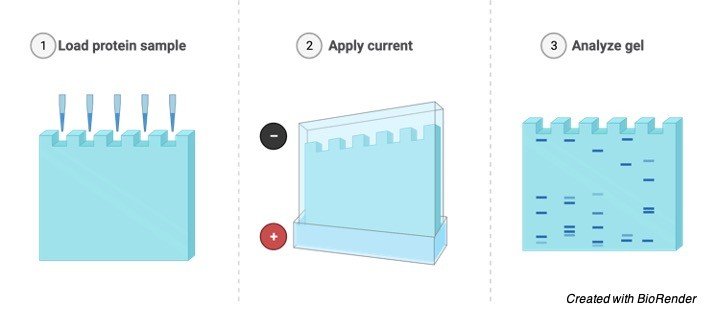
Vertical Gel electrophoresis
The rate of Migration of molecules in an electric field can be calculated according to the size and number of groups of charged particles in one molecule, Electrophoresis is used for the separation of proteins, amino acids and nucleic acids.
Application of Gel Electrophoresis
Gel electrophoresis is used for the separation of proteins such as DNA, RNA and other macromolecules depending on their size and charge of the particles.
As mentioned above it is technique where the gel containing the molecules is allowed to run on the electric field where current is passed through and the molecules will be separated depending on the size and charge by passing at different directions with different speed which helps them to separate accordingly.
Considering all the molecules have the same amount of charge so it can be separated by gel electrophoresis by considering their size.
Now let us know how the DNA fragments are separated even though they have the same size relatively depending upon their fragments.
We can measure the absolute size of the DNA by comparing it with the standard yard stick which is being made up of known sizes of DNA fragments,
Principle of Gel Electrophoresis
This technique is generally known as gel electrophoresis because the supporting film is made up of jelly like substance which is made up of Polysaccharide named agarose.
It is usually in a powdered dry form as flakes and it can be converted into a gel form by heating in a buffer solution containing water and some kind of salts and further it is allowed to cool so that it forms like a solid slimy gel.
When considering the gel at molecular level it is seen that the agarose molecules present in it are held together by hydrogen bonds with minute pores in it.
At the end of the supporting film two well like depressions are seen where the samples are kept to undergo the test.
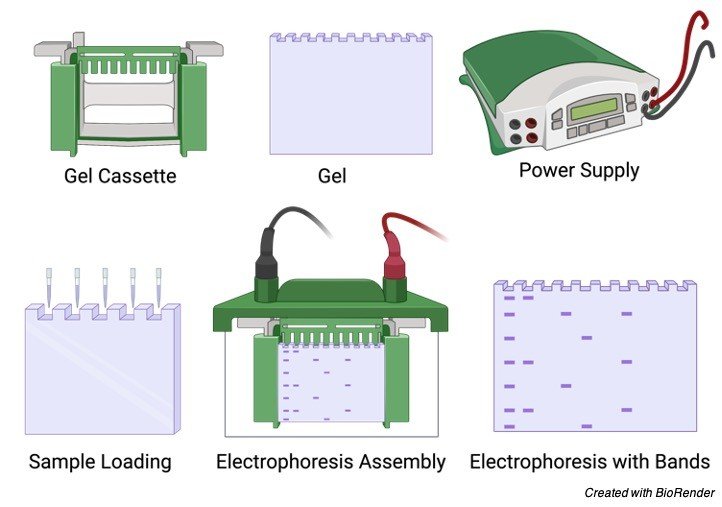
The prepared gel is then placed in a gel box and the samples are added or kept in the well and the positive electrodes are placed at one end of the gel box and the negative electrode in the opposite direction.
Then the salt solution which helps in conducting the current is poured on the gel which is being placed in a gel box.
This forms a glassy layer over the gel. The end of the box where wells are present is marked as negative electrode and the other side without wells is considered as positive electrode.
Gel Electrophoresis Samples
Once the gel is placed in the gel box, the samples are being loaded in the wells, one well is kept consigned for DNA ladder, which is the known length of DNA fragments, these ladders can be purchased commercials according to our size and needs.
Considering an example of PCR reaction here, after loading the DNA ladder, in one well, the samples needed for the pcr reaction is loaded in the remaining wells and the gel box is switched on and the current is allowed to flow through the gel box.
As the DNA molecules contain a sugar phosphate group, they carry a negative charge and started moving towards the positive charge.
Maximum of 80 to 120 V is used to run the gel, more than this high voltage is not recommended as it allows the gel to melt even though it coordinates in faster moving.
Once the gel is allowed to run the smaller DNA fragments flows faster while the larger ones does not move and stays near the wells.
Visualization
Sample (DNA, RNA or Proteins) fragments are thus separated by size of the particles accordingly.
Small particles are found at the end of the gel and larger particles near the wells.
To examine the size of the bands that are found on it, this process is being carried out by staining the gel with staining dye for e.g. DNA-Staining dye and by placing it under the uv light.
The bands start glowing and thus allowing us to see the location of the DNA throughout the gel.
Band mentioned here is nothing but a well-defined line that runs along the DNA, where each band contains large number of fragments of DNA with same sizes.
A band is necessary for the visualization of DNA because they are not visibly seen as such lying on the gel.
Thus, the band helps us to determine the size of their fragments depending upon their location on the band.
Gel Electrophoresis Citations
- Understanding gel electrophoresis. Nature . 1990 May 31;345(6274):381.
- Introduction to Protein Electrophoresis. Methods Mol Biol . 2019;1855:23-29.
- High Sensitivity Protein Gel Electrophoresis Label Compatible with Mass-Spectrometry. Biosensors (Basel) . 2020 Oct 31;10(11):160.
- Two-dimensional gel electrophoresis. Curr Protoc Immunol . 2005 Nov;Chapter 8:Unit 8.5.
- Ultrarapid Sodium Dodecyl Sulfate Polyacrylamide Mini-Gel Electrophoresis. Methods Mol Biol . 2019;1855:491-494.
Share
Similar Post:
-
Gene Regulation: Definition, Steps, and Examples
Continue ReadingWhat is Gene Regulation?
Have you all ever been amazed at how our body makes different types of cells and how those cells perform different functions?
Generally, our body is made up of hundred types of cells which are different in their forms and vary with their functions, accounting from our skin to our immune and blood cells.
All these cells contain the same genetic material, ie. DNA but these cells vary in their structure and function, because of gene regulation.
Gene regulation is one of the processes in which the genes are turned on and off. During the early development of an organism, the cells beings to take up specific mechanisms.
The Gene regulation process helps in ensuring, whether the genes are expressed in an appropriate manner and at a proper time.
Gene regulation helps an organism to respond to environmental conditions.
Gene regulation is activated by various mechanisms like modification of genes chemically, use of regulatory proteins to switch on and off the genes, etc.
The human genome consists of almost 20,000 genes. Out of which 10, 000 are actively present in the cells, and the remaining 10,000 will be in an inactivated form.
Such that the cell determines which gene will be active and which will not be activated, ie. In the inactive form.
Gene Regulation Features
Gene regulation is a mechanism that controls the genes by activating a few of them out of the many genes that are present in its genomes.
Gene regulation is very much important as each cell in our body has a different set of genes, interestingly, all cells in our body have the same DNA, but these differ in their expression of the genes, causes variations among them and each of the cells has a different set of proteins which makes them unique in their function.
One such example of gene regulation is the functioning of the liver. The function of the liver is to remove the toxic substances that are present in our body such as alcohol and chemical agents in our bloodstream that we intake through some kinds of food substances.

To make the liver function, liver cells express a gene encoding subunits in an enzyme which is known as alcohol dehydrogenase.
This enzyme breaks the alcohol into a non-toxic molar substance. Then the neurons in the brain get activated and send particular signals to remove the toxins from the body of an individual, at this time the genes are unexpressed, which means that they are turned off, in the same way when the genes of the liver cells are in functions, the genes of the neurotransmitters are turned off.
How Does Gene Regulation Work?
As mentioned above, different cells express different types of gene regulations. In some cases, two different types of cells of the same kind have different types of expressions based on environmental conditions.
However, the gene expression pattern is determined from both inside and outside the cell. Where the example that can be sorted for gene expression from inside a cell is the inheritance of proteins from its mother cell in case of any damage in the DNA and the amount of ATP it has.
The example that can be sorted for gene expression from outside the cell is that the chemical signals which are arising from the other cells in the form of mechanical signals from the extracellular matrix and the nutrient levels in the cells.
How Do Cells Control Gene Regulation?
First of all, cells do not make decisions that, which cell has to generate an expression. Instead of the cells, undergoes molecular pathways which help them to converts the information in such a way that it binds to its specific receptors with the help of a chemical signal that is generated as a result of gene expression.
Let us consider an example of how cells react to the growth factors. The growth factor is one of a chemical signal which arises from a neighboring cell to instruct a target cell to grow and divide.
This process occurs in such a way that the cell detects the growth factor by binding to the growth factor physically which is present on a cell surface as a receptor protein.
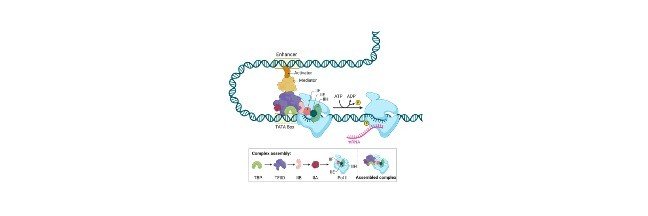
Such binding of a growth factor causes the receptors to change their shape, by triggering a series of chemical events in a cell that activates proteins, and it is known as transcription factors.
Further, these transcription factors bind to a certain sequence of the DNA that is present inside a nucleus and causes transcription of the genes to activate cell division.
As a result, the genes which is the product of different proteins make the cells divide and induce growth in a cell.
Eukaryotic Gene Regulation
Gene regulation is a result of various kinds of expressions in a gene. Let us discuss how the genes are being expressed in various stages depending on the availability of the DNA to produce the mRNAs which help in translating and processing the respective proteins.
This Eukaryotic gene expression involves many steps and all these can be regulated. Different genes are being regulated at different intervals.
The expression of eukaryotic genes involves many factors such as follows.
I. Chromatin Accessibility
The structure of chromatin is very much important for the regulation of the genes as the more open or relaxed the chromatin makes the gene more available for transcription.

II. Transcription
Transcription acts as a key for the regulation of many genes. A set of transcription factors helps a protein to bind with a specific sequence of DNA in or near a gene which either promotes or represses a transcription into an RNA.
III. RNA Processing
RNA processing involves splicing, capping, and an addition of a poly-A tail to regulate a molecule of RNA. Various mRNAs are made from the same pre-RNA by the process of alternative splicing.
IV. RNA Stability
The lifetime of an mRNA molecule in a cytosol affects many proteins which are made from it. Many of the small regulatory RNAs can bind to the target mRNAs which causes them to chopped up Whereas the small regulatory RNAs are called miRNAs.
V. Translation
Translation of an mRNA can be increased by the regulators. Protein activity: Proteins undergo a variety of modifications, which are being chopped up or tagged with the chemical groups.
These modifications can be regulated which also affects the activity or behavior of the proteins.
Although all the stages of gene expression are regulated, the main control activity of the gene is transcription.
Further process of gene regulation refines the patterns of gene expression that are roughed out during transcription.
Gene Regulation Citations
- RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem Sci . 2015 May;40(5):248-56.
- The Evolution of Gene Expression in cis and trans. Trends Genet . 2018 Jul;34(7):532-544.
- Enhancer DNA methylation: implications for gene regulation. Essays Biochem . 2019 Dec 20;63(6):707-715.
- Gene Regulation in and out of Equilibrium. Annu Rev Biophys . 2020 May 6;49:199-226.
- Epigenetic regulation and chromatin remodeling in learning and memory. Exp Mol Med . 2017 Jan 13;49(1):e281.
- Building Transcription Factor Binding Site Models to Understand Gene Regulation in Plants. Mol Plant . 2019 Jun 3;12(6):743-763.
- Crossroads between transposons and gene regulation. Philos Trans R Soc Lond B Biol Sci . 2020 Mar 30;375(1795):20190330.
- Fighting the Fire: Mechanisms of Inflammatory Gene Regulation by the Glucocorticoid Receptor. Front Immunol . 2019 Aug 7;10:1859.
- The role of 3D chromatin domains in gene regulation: a multi-facetted view on genome organization. Curr Opin Genet Dev . 2020 Apr;61:1-8.
Share
Similar Post:
-

What is DNA Fingerprinting? Steps, Definition, Application,...
Continue ReadingWhat is DNA Fingerprinting?
DNA fingerprinting is a technique used in genetics for isolating and identifying the variable elements in the sequence of the base pair in the DNA.
DNA fingerprinting is also called out in many forms as DNA profiling, genetic fingerprinting, genotyping or as identity testing.
This technique was first developed by British Genetist Alec Jeffreys in the year 1984 when he was noticing the certain sequences in a highly variable DNA, which is also known as minisatellites, that do not contribute the functions of the genes and the genes are repeatedly present.
He also identified that each one has a unique pattern of the minisatellites.
Features of DNA Fingerprinting
The DNA Fingerprinting method used to identify the identity of an individual apart from his physical characteristics with the sample of DNA by identifying the unique patterns in it is known as DNA fingerprinting.
We all know that each and every cell, in our body contains DNA. Nearly 99.9 percent of the DNA between the humans is similar, where as the remaining one person makes each of us a unique individual, and this probability reduces down in case of identical twins.
Though This unique remains as a small amount there are about three million base pairs are differed between the peoples.
Minisatellites are the short sequence consisting of about 10 to 60 base pairs of the repetitive DNA which shows a greater variation from one person to the other and even in other parts of a genome. The minisatellite was first discovered in the year 1980.
DNA fingerprinting detects lots of minisatellites in the genome of an individual and thud helps us in finding the uniqueness between the two individuals or the difference.
As we all have our own fingerprinting, DNA fingerprinting is also unique with which we are born with.
DNA Fingerprinting Steps
Collection of an organic sample such as blood, buccal swap, spit, semen or a tissue.
DNA is extracted from the samples.
Restricted absorption or PCR intensification takes place.
Electrophoresis is performed
Uninterpreting outcomes are collected.
DNA Fingerprinting Process
The process followed for the DNA finger printing are discussed as follows;
Sample Collection: DNA is acquired from the body samples either in the form of liquids such as blood or semen, saliva, amniotic fluid, etc. or the tissues from particular parts.
DNA Extraction: For the process of DNA fingerprinting extracting the DNA is very much essential. High quality and the good amount of DNA is needed as it expands the conceivable outcomes for getting better results.
DNA extraction stragedy can be performed by the following strategy;
• Phenol-chloroform DNA strategy
• CTAB DNA extraction strategy
• Proteinase K DNA extraction strategy.
The immaculateness and the quantity of DNA needed could be of about 1.80 and 100ng, individually to undergo a DNA test.
DNA is filtered using the DNA sanitation in it, if it is necessary. Then the DNA is measured using the UV visible spectrophotometer and it performs one of the accompanying strategies recorded underneath.
Restriction, Absorption, Enhancement
Restriction, enhancement, absorption or DNA sequencing is performed by following the regular tragedies as
• RFLP based STR investigation
• PCR based investigation
• Real-time PCR investigation.
DNA Fingerprinting Result Analysis
After examining under a southern blotting agarose gel electrophoresis, narrow electrophoresis, ongoing intensification and the DNA sequencing the outcomes for the different DNA profiles are compared with the RT-PCR sequencing. This method is used widely in the forensic science.
Interpreting DNA Fingerprinting Results
By over viewing the DNA profiles, of different individuals, varieties, likeness and similarities between different individuals are distinguishes. Now a days all these procedures are in an automatic manner and it is computerised and we are very much thankful to the technologies.
Application of DNA Fingerprinting
Using the technique of DNA fingerprinting, the natural personality of an individual can be identified. For the identifying the ones character there is no need for alternative preferable samples of the DNA.
Gravely eradicated dead bodies can be identified.
DNA fingerprinting helps in detecting the maternal contamination of a cell.
One of the significant uses of this technique is to determine the maternal cell tainting. The amniotic fluid or the CVS test contains the maternal DNA once in a while. Contamination of the sample have the great chances of giving the false statements particularly in carrier recognition.
Using VNTRs and the STRs markers with the PCR gel electrophoresis, maternal cell tainting can be identified during the time of pregnancy hereditary testing.
The most significant use of this technique in a present-day generation is to find a crime report or a criminal check.
The sample that is taken from the crime site like salivation, blood, hair follicle or semen is collected and the DNA from the particular sample is separated and it is investigated against the suspect by utilizing the two markers which helps us to clarify.
By coordinating the band design of the DNA, criminals connected to it can be found.
DNA Fingerprinting Citations
- Age Estimation with DNA: From Forensic DNA Fingerprinting to Forensic (Epi)Genomics: A Mini-Review. Gerontology . 2018;64(4):326-332.
- DNA fingerprinting in the criminal justice system: an overview. DNA Cell Biol . 2006 Mar;25(3):181-8.
- DNA fingerprinting, DNA barcoding, and next generation sequencing technology in plants. Methods Mol Biol . 2012;862:13-22.
Share
Similar Post:
-

Lac Operon: Diagram, Phenotypes, Model, and Regulation
Continue ReadingLac Operon
Lac operon is an abbreviated form of lactose operon. Lactose operon is used for transporting and metabolising the lactose in E. coli and many other enteric bacteria.
Though glucose is considered a preferred carbon source for most bacteria, the activity of operon allows for the digestion of lactose effectively when the glucose is not available through the activity of the beta-galactosidase.
Gene regulation of the lac operon is considered as the first genetic regulatory mechanism which helps us to understand the concept clearly.
So, it became a common example of prokaryotic gene regulation. Before knowing about the lac operon, it is very much important to know about gene regulation.
Features of Lac Operon
Lac operon is generally defined as the group of genes which has a single promoter that encodes the genes for the transport and the metabolism of the lactose in the bacterium Escherichia coli and other species of bacteria.
Gene regulation in the prokaryotes can be explained with the help of the model of the lac operon.
The alteration in the physiological and environmental conditions are observed leading to an alteration in the expression of genes in a prokaryote.
This concept was first observed by Jacob and Monod.
The lac operon consists of the following;
Regulatory gene: It is usually represented by the letter I, It codes for the repressor proteins.
z gene: Its codes for the enzyme beta-galactosidase which catalyzes the hydrolysis of the lactose into glucose and galactose.
Y gene: This gene codes for the enzyme permease which helps in regulating the lactose permeability in the cell.
a gene: It usually codes for transacetylase and assists in the formation of the enzyme beta-galactosidase.
All these genes help in lactose metabolism. However, In the lac operon, lactose acts as an inducer.
The regulatory gene is activated when the lactose is provided with a medium of bacteria. Then the lactose which is acting as an inducer binds with the repressor proteins and renders the protein to get inactivated and allows the transcription of the operon. So, the lac operon is negatively regulated in such cases.
Bacterial Lac Operon
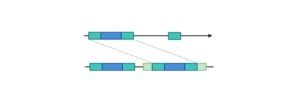
Bacterial operons are polycistronic transcripts, that can produce multiple proteins from a single mRNA transcript. When lactose is required as a sugar source for the bacterium, three genes of the lac operon are expressed, and also their subsequent proteins are translated: lacZ, lacy, and lac A.
Whereas the gene product of lac Z is known as beta-galactosidase which plays an active role in cleaving lactose into a monosaccharide as glucose and galactose.
Lac Y encodes for the enzyme Beta-galactosidase permease which is a membrane protein and it becomes embedded in a cytoplasmic membrane and thus enables the cellular transport of the lactose into the cell and lac A encodes for Galactosidase acetyltransferase; this is used to produce the enzymes when there is no availability of the lactose, it has also been used when enough glucose is not available to get a required amount of energy.

Lac operon generally uses two parts as the control mechanism for ensuring the energy expenditure by the cells for the production of enzymes that are encoded by the lac operon only when they are necessary.
At the time of absence of lactose, lac repressor (lacI) depresses or halts the production of enzymes that are encoded by lac operon.
Lac repressor is always in an expressed form unless a co-inducer binds to it. It can also be said that lac operon transcribes only in the presence of a small molecule co-inducer.
In the presence of glucose, catabolite activator protein is required for the production of enzymes, which shutdowns lactose permease for preventing the transport of lactose in a cell.
This dual mechanism causes the sequential utilization of glucose and lactose into two distinct phases of growth, which is known as diauxie.
Structure of Lac Operon
Lac operon contains about 3 structural genes along with a promoter, regulator, terminator, and operator. The structural genes are named lacz, lacY, and lacA.
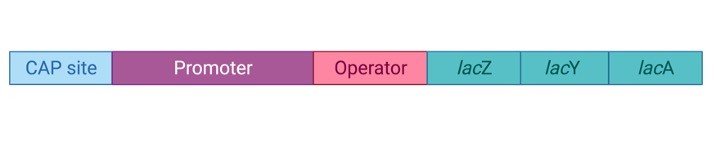
Lac Z encodes for Beta-galactosidase, which is an intracellular enzyme and cleaves the disaccharide lactose into glucose and galactose.
Lac Y encodes for Beta-galactoside permease, which is a transmembrane symporter that pumps the Beta-galactosides including lactose into the cell using a gradient of protein in the same direction.
Permease increases the permeability of the cell. Lac A encodes for Beta-galactosidase transacetylase which transfers an acetyl group, acetyl-CoA to thiogalactoside.
In this only lacZ and lac, Y is needed for the catabolism of the lactose.
Regulation of Lac Operon
The activity which controls the expression of the lac operon is regulated by two different kinds of proteins. Where one of the proteins prevents the RNA polymerase from transcribing and is known as the negative control, and the other protein enhances the building of the enzyme RNA polymerase and is known as the positive control.
Negative Regulation of Lac Operon
The proteins which inhibit the process of transcription in lac operon is a tetramer with four identical subunits and they are generally known as lac repressor.
This lac repressor is encoded by a lacI gene, which is located upstream of the lac operon and also consists of its promoter.
Expression of lacI gene is not regulated and only very low levels of the lac repressor are synthesized continuously.
Genes whose expression cannot be regulated are said to be constitutive genes. At the time of absence of the lactose, the lac repressor blocks the expression of the lac operon which is downstream of the promoter and upstream of the transcriptional initiation site.
The operator also consists of a specific nucleotide that is recognized by the repressors which bind tightly and block the transcription.
The lac repressors have a very high affinity for the lactose, when a small amount of lactose is present, lacI binds and dissociated from the DNA operator, and frees operons from regulating the gene expression.
Such substances which cause repressors to dissociate from operators are known as inducers and the genes regulated by the repressors are known as inducible genes.
Positive Regulation of Lac Operon
Though lactose induces the expression of the lac operon, the expression level is very low, the reason for this is due to the reduced expression of the genes and it can be regulated with the presence of glucose.
Glucose is easily metabolized so it is prefeed as a source over lactose to prevent the expression of the lac operon.
The promoter for the lac operon is weak and it is poorly transcribed for induction. The binding site is present upstream from the promoter, for a protein known as CAP (catabolic activator protein).
When CAP binds it distorts the DNA which results in RNA polymerase binding in an effective manner and transcription is very much enhanced in the lac operon.
To bind the CAP must bind initially with the cAMP which increases the activating CAP and stimulates the transcription.
Thus, the cAMP and the CAP complex are referred to as positive regulators.
Lac Operon Citations
- Modeling network dynamics: the lac operon, a case study. J Cell Biol . 2003 May 12;161(3):471-6. 4;16(4):1729-1749.
- Analytical Expressions and Physics for Single-Cell mRNA Distributions of the lac Operon of E. coli. Biophys J . 2019 Aug 6;117(3):572-586.
- Stability and Hopf bifurcation analysis of lac Operon model with distributed delay and nonlinear degradation rate. Math Med Biol . 2019 Dec 4;36(4):489-512.
- Kinetic approaches to lactose operon induction and bimodality. J Theor Biol . 2013 May 21;325:62-75.
- Weak operator binding enhances simulated Lac repressor-mediated DNA looping. Biopolymers . 2013 Dec;99(12):1070-81.
- Bistability and Asynchrony in a Boolean Model of the L-arabinose Operon in Escherichia coli. Bull Math Biol . 2017 Aug;79(8):1778-1795.
- Induction of the lac promoter in the absence of DNA loops and the stoichiometry of induction. Nucleic Acids Res . 2006 Jan 23;34(2):606-12.
- Effect of lac repressor oligomerization on regulatory outcome. Mol Microbiol . 1992 Apr;6(8):963-8.
Share
Similar Post:
-

Genetic Code: Chart, Table, Definition, and Examples
Continue ReadingWhat is Genetic Code?
Genetic code is one of the set of rules which is used by all the cell in the living organisms to translate the information which is encoded within the genetic material as DNA or mRNA which is the sequences of the nucleotides triplet or codons which make up the proteins.
This translation is accomplished by the cell organelle named as ribosomes, which connects the proteinogenic amino acids in order that is specified by the mRNA, with the help of transfer RNA which carries the amino acids and make it to read by the mRNA three nucleotides at the specific time.
The genetic code is similar to all organism and it is expressed within the cells in a simple way.
This codon specifies the amino acid which will be added to the next during protein synthesis. However, with some of the expectations, three nucleotide codons in a nucleic acid sequence specifies a particular amino acid.
Majority of the genes are encoded with the appropriate way according to its function. The observation of how proteins are encoded begins after the discovery of structure in year 1953.
George Gamow postulated the three bases which are needed to encode the 20 amino acids which are used by the living cells that are used to build up the proteins. It allows a maximum of 64 amino acids.
Features of Genetic Code
Genetic code is defined as the sequence of nucleotides present in the nucleic acids such as DNA (Deoxyribonucleic acids) or the RNA (Ribonucleic acids) which determines the amino acid sequence of the proteins.
Even the linear sequence of nucleotides in the DNA contains the information for the protein sequences, where proteins are not made directly from the DNA, rather the linear sequence of the nucleotides in the DNA contains specific information for the protein sequences, Proteins are not synthesized directly from the DNA and which directs the formation of the protein.
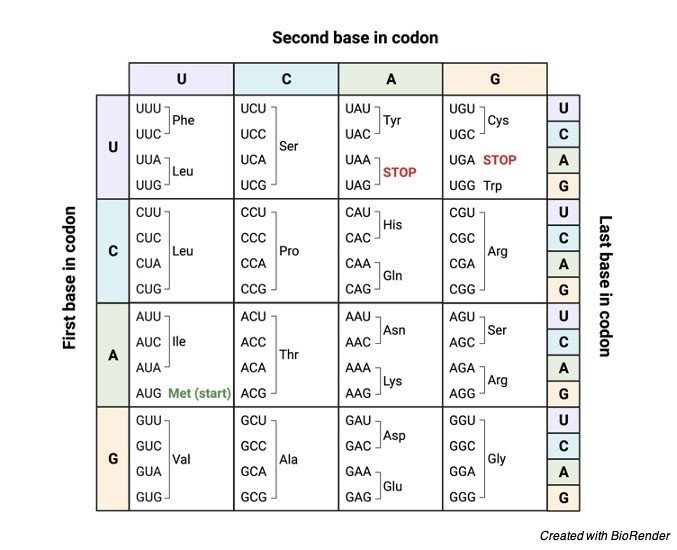
Genetic Code Chart
RNA is also composed of four nucleotides namely adenine, guanine, Cytosine and Uracil. Where as the three adjacent nucleotides constitute a unit known as the codon, which codes for the amino acid.
For example: the sequence of the codon AUG which specifies the methionine amino acid.
There are almost 64 possible codons, out of which three does not code for amino acids, but these indicates the end of the protein.
The remaining 61 codons specifies the 20 amino acids which make up the proteins the codon AUG additionally indicates the start of the protein along with the coding of the methionine.
Methionine and tryptophan are considered as the two amino acids which are coded by just a single codons AUG and UGG respectively.
The remaining 18 amino acids are coded by the two to six codons. The amino acids are coded by more than a single codon and it is called as a codon-degenerate.
The genetic code once thought to be identical in all forms of life, which has been found diversely in certain organisms and also in mitochondria in some eukaryotes.
Types of Genetic Code
The genetic code is differentiated into two types and they can be expressed in the form of either RNA codons which occur in the messenger RNA and the codons reads the information during the production of polypeptides during the process of translation.
Each of the messenger RNA molecules acquires their sequence of nucleotides by the process of transcription from the corresponding DNA.
Because during the process, DNA sequencing becomes rapid because most of the genes are now being discovered during the level of DNA before they are discovered as mRNA or as a product of protein, which is extremely useful to have the table of codons which is expressed as DNA.
DNA Codons
These codons are read on the sense strand of the DNA. Except the nucleotide Thymine(T) which is found in the place of Uracil, which reads the same as RNA codons. mRNA is synthesized with the help of antisense strand of the DNA which acts as a template.
Types of Codon
The genetic code has about 64 triplets of nucleotides. The triplets are known as codons, with the three expectations, each of the codon encodes for one of the 20 amino acids for the production of proteins.
This produces some of the redundancy in code. Almost all the amino acids are encoded more than a single codon.
Where one codon is AUG which serves the two related functions. It also signals for the start of translation, and its codes for the incorporating of the amino acid methionine into the growing chain of the polypeptide chain.
The codons are of two types namely;
Sense codons
Signal codons
I. Sense Codon
Sense codons codes for the amino acids. There are about sixty-one sense codons in the genetic code, which has to be code for 20 amino acids.
II. Signal Codon
Sense codon codes for the signaling during the production of proteins. There are about four codons that codes for a signal, namely AUG, UAA, UAG and UGA. Signal codons are further classified as two types namely
Start codons: The codon which starts the process of translation is known as start codon. It is also commonly called as initiation codons as it initiates the production of polypeptide chain. The best said example for start codon is AUG. This codes for the amino acid methionine, in prokaryotes it is N-formyl methionine.
Stop codons: Stop codon provides a signal for termination of a polypeptide chain. These codons are commonly called as termination codons as they provide a signal for the termination and the release of the polypeptide chain.
Properties of Genetic Code
Genetic code-Triplet: It consists of 64 codons, which are sufficient to code the 20 amnio acids and it helps in signaling the start and stop codons and synthesizing the polypeptide chain. Where as a in a triplet code 3 bases of RNA codes for a single amino acid.
The code is also universal which are assigned for the same amino acids.
The genetic code is comma less where the codons are continuous and are demarcation lines between the codons.
The genetic code is usually non-over lapping. Whereas in non-overlapping two amino acids are being coded by the six bases.
The genetic code is also non-ambiguous, Redundant and it has a polarity.
Genetic Code Citations
- The evolution of the genetic code: Impasses and challenges. Biosystems . 2018 Feb;164:217-225.
- The Cipher of the Genetic Code. Biosystems . 2018 Sep;171:31-47.
- Origin and Evolution of the Universal Genetic Code. Annu Rev Genet . 2017 Nov 27;51:45-62.
- The path to the genetic code. Biochim Biophys Acta Gen Subj . 2017 Nov;1861(11 Pt A):2674-2679.
- Understanding the Genetic Code. J Bacteriol . 2019 Jul 10;201(15):e00091-19.
- Rewriting the Genetic Code. Annu Rev Microbiol . 2017 Sep 8;71:557-577.
Share
Similar Post:
-

Calvin Cycle: Diagram, Definition, and Mechanism
Continue ReadingCalvin Cycle
Plants are indispensable in world sustaining life forms in earth; producing basic food components from inorganic compounds: CO2, HCO3, etc.
The conversion from inorganic to organic compounds is a light mediated photosynthetic process and a characteristic feature of plants; where they synthesis food using light are called as Autotrophs.
Atmospheric CO2 present as a mixture of gases – Oxygen and Nitrogen; in the stratosphere; formed as a final product of respiration and man.
Carbon is a principle inorganic molecule constituting a skeleton for all organic compounds. One can say that carbon skeleton is basis for all living organism and it circulates from organism to organism then in atmosphere are fixed again in food components by plants enters organism that consumes it.
CO2 fixation is a major process in plants where they utilize atmospheric CO2 to synthesize sugar molecules (i.e.) Starch in the presence of light (photosynthesis).
Starch is stored in plants; plant products are consumed by herbivores and then it enters the food chain involves in structural and physiological functions thereby maintaining homeostasis by participating in various metabolic pathways; as an end product the CO2 is again released to the atmosphere.
Photosynthesis is engrossed in oxygen production and CO2 assimilation at two stages in chloroplasts.
Water oxidation yields oxygen in thylakoid and carbon dioxide reduction to carbohydrates in stroma of chloroplast; powered by ATP and NADPH produced from the oxidation.
Second part of photosynthesis reducing carbon dioxide; is light independent after the enzyme mediating the reaction is activated; called as dark reaction or Calvin Cycle.
Biological CO2 fixation is a significant feature of plants and certain algae and chemoautotrophs.
Alternative Names of Calvin Cycle
Photosynthetic Carbon Reduction Cycle (PCR Cycle), Calvin – Benson Cycle, Reductive Pentose Phosphate Pathway and C3 cycle.
Discovery of Calvin Cycle
Calvin Cycle was discovered by a group of 3 members: Melvin Calvin, Andrew Benson and James Bassham in Berkley deduced the cycle between 1940 to 1953 using C – 14 radioactive isotope which were abundant during the period.
The organism was Chlorella, an algal autotrophic colony. The process is rapid to produce Glyceraldehyde 3 phosphate an inter convertible triose phosphate and regenerates a chemical component taken up in chloroplasts.
Mechanism of Calvin Cycle
Carbon dioxide bio fixation is done in Calvin Cycle in 13 reactions catalyzed by 11 enzymes in stroma. The cycle has 3 phases where the carbon dioxide is reduced and Ribulose – 1, 5 – Bisphosphate is regenerated. Ribulose – 1, 5 – Bisphosphate acts as a primary skeleton to carry CO2 and produce starch.
Three molecules of carbon dioxide are utilized to produce single triose phosphate (i.e.) DHAP.
Phases of Calvin Cycle
The 3 phases are:
(i) Carboxylation: Atmospheric CO2 is fixed by a pentose sugar Ribulose – 1, 5 – Bisphosphate to form 2 molecules of 3 – Phosphoglycerate.
(ii) Reduction: The reaction involves the reduction of 3 – Phosphoglycerate is converted into Hexose sugar molecules when it enters gluconeogenesis.
(iii) Regeneration: The involved Ribulose – 1, 5 – Bisphosphate to regenerate which can be used for many products.
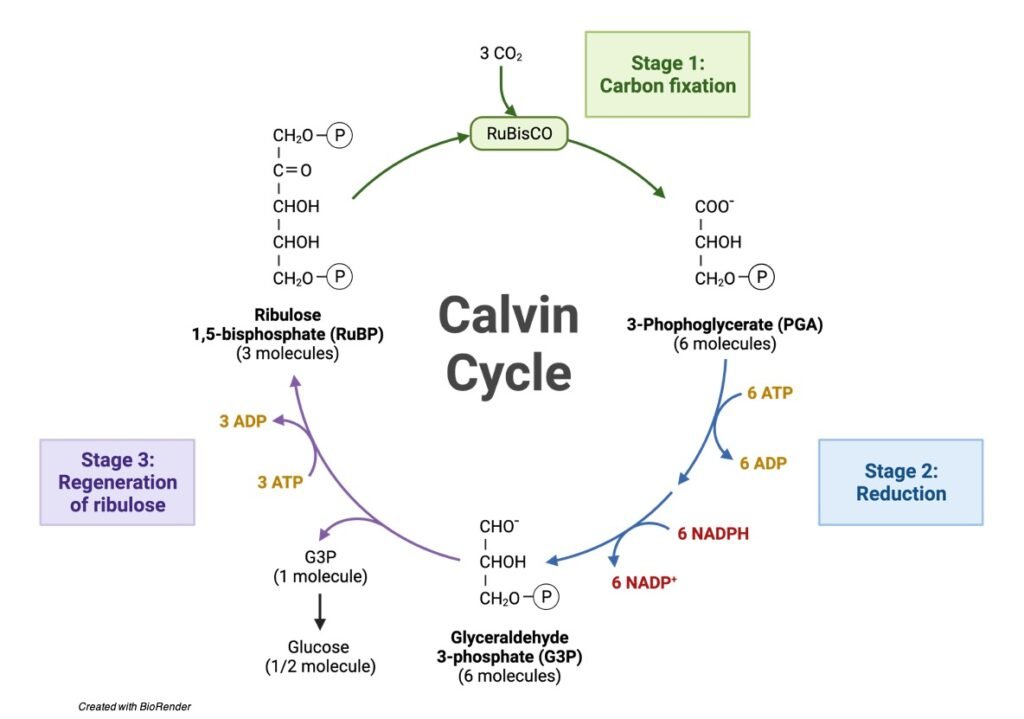
(i) Carboxylation
The initial phase of carbon fixation reaction utilizes carbon dioxide. R – 1, 5 – BP will be attached to Mg+ ion of Ribulose – 1, 5 – Bisphosphate carboxylase/ oxygenase (Rubisco) enzyme and forms an intermediate – Enediolate intermediate, a deprotonated form.
This deprotonated form facilitates CO2 attachment in 4th carbon also, an intermediate of ketoacid. Water molecules enters the reaction to hydrate the pentose intermediate at keto group.
Hydrated pentose sugar yields two 3 – Phosphoglycerate (3 – PGA) molecules. This mechanism is possible when Rubisco containing both carboxylase and oxygenase acts as a carboxylase fixing CO2 to the Pentose sugar.
Three molecules of CO2 react with 3 molecules of R – 1,5 – BP to produce six 3 – PGA molecules.
(ii) Reduction
ATP and Reduced equivalents reduce the C3 carbon to C6 carbon sugars. The reaction converts six 3 – PGA molecules into 6 molecules of Glyceraldehyde – 3 – Phosphate.
Enzyme: 3 – phosphoglycerate kinase and NADP – Glyceraldehyde – 3 – Phosphate Dehydrogenase.
3 – phosphoglycerate kinase catalyzes the conversion of 3 – PGA into 1, 3 – Bisphosphoglycerate by adding a phosphate group from ATP to 1st carbon atom of the molecule.
Similar to gluconeogenetic pathway 1, 3 – Bisphosphoglycerate is converted Glyceraldehyde – 3 Phosphate utilizes reduced equivalent NADPH to form Glyceraldehyde – 3 – Phosphate removing 6 phosphate groups from six molecules of 1, 3 – Bisphosphoglycerate.
The GAP can be transported to cytosol for Carbohydrate compounds in its interconvertible form DHAP.
(iii) Regeneration
Step1: The Primary CO2 fixator molecule yields, a molecule of Hexose sugar which comprises the 3 CO2 molecule absorbed from the atmosphere. The other 5 GAP undergoes further reaction to regenerate 3 Ribulose – 1,5 – Bisphosphate.
Step2: Two molecules of DHAP are formed by the catalyzes of Triose phosphate isomerase. This is the initial step where it forms a reversible DHAP for the formation of Ribulose – 1,5 – Bisphosphate.
Step3: Enzyme Aldolase will condense a molecule of DHAP and GAP to form Fructose – 1,6 – Bisphosphate.
Step4: Fructose 1, 6 – bisphosphatase facilitates the hydration of the F – 1,6 – P to convert to Fructose – 6 – Phosphate.
Step5: the 4th molecule of GAP receives two carbons from F – 6 – P and catalyzed by Transketolase. The product formed is Erythrose – 4 – Phosphate and Xylulose 5 – Phosphate.
Step6: Erythrose – 4 – Phosphate combines with last DHAP molecule to produce 7 Carbon compound Sedoheptulose – 1, 7 – Bisphosphatase mediated by Aldolase.
Step7: Sedoheptulose – 1, 7 – Bisphosphatase is hydrolyzed to form Sedoheptulose – 7 – Phosphate.
Step8: Transketolases transfers two carbon groups of Sedoheptulose to GAP forming Ribose – 5 – Phosphate and Xylulose – 5 – Phosphate.
Step 9 & 10: Xylulose – 5 – phosphate and Ribose – 5 – Phosphate is converted into Ribulose – 5 – Phosphate by the enzymes Ribulose – 5 – Phosphate epimerase and Ribose – 5 – Phosphate isomerase respectively.
Step11: R – 5 – P in the presence of enzyme Phosphoribulokinase converts into 3 molecules of R – 1,5 – BP.
Features of Calvin Cycle
The Calvin Cycle is dependent on few environmental conditions. They are: pH, Mg, NADPH and reduced ferredoxin concentration along with Light.
1. Rubisco enzyme is activated by light. Higher the amount of radiance increased rate of enzyme activity.
2. Light increases the pH level by favoring carbamate formation and Mg
3. Thioredoxin regulates the Calvin cycle by activating enzymes which are used in synthesis of organic compound and degrades the enzymes which inhibit the synthesis.
Calvin Cycle Citations
- Photosynthesis: basics, history and modelling. Ann Bot . 2020 Sep 14;126(4):511-537.
- Kinetic modeling of the Calvin cycle identifies flux control and stable metabolomes in Synechocystis carbon fixation. J Exp Bot . 2019 Feb 5;70(3):973-983.
- Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev Mar Sci . 2011;3:261-89.
- The minimal model of Hahn for the Calvin cycle. Math Biosci Eng . 2019 Mar 15;16(4):2353-2370.
- The importance of the photosynthetic Gibbs effect in the elucidation of the Calvin-Benson-Bassham cycle. Biochem Soc Trans . 2018 Feb 19;46(1):131-140.
- Discovery of the canonical Calvin-Benson cycle. Photosynth Res . 2019 May;140(2):235-252.
Share
Similar Post:
-

Seed Dormancy: Germination, Definition, and Mechanism
Continue ReadingSeed Dormancy
Optimal conditions support the seed germination under prerequisite condition; hydration of seed marks germination of the plant.
The absence or minimal expression of prerequisite conditions will make the seed to enter a stage of inactivity – quiescence stage.
The metabolic conditions are pertained in this state but the differentiation and development are inhibited. The state is termed as Dormant stage.
Dormancy is a stage of inactivity induced in developing plant parts when subjected to stress such as insufficient water imbibition, low temperature, oxygen or other factors; establishing an undesirable effect on plant germination simultaneously increasing the survival capacity of the plant by temporary impairment in development of plants.
Seed dormancy is an inherited phenomenon acquired during the course of evolution in almost all species of plants.
The phenomenon remains untraceable through the evolutionary history of plants. But a major understanding is that, plants in equatorial region show reduced or no dormancy.
As we move away from the equator the plants increase the tendency of dormancy. Though many factors are involved in determining the dormancy a true working mechanism of dormancy yet remains unclear.
Understanding seed dormancy prevailed from 372 BC, Greek philosopher Theophrastus known as the Father of Seed Physiology has studied seed dormancy.
Recent studies on seed dormancy are minimal and the mechanism and factors involved are not discovered authentically.
Characteristic Features of Seed Dormancy
1. An acquired inherited feature: The process is an age-old characteristic feature of plants which are subjected to constantly varying environment is acquired and passed on from generation to generation.
2. Inactivation is temporary: The dormant seed remains the same till a favorable condition develops to support the germination.
3. Defense mechanism: The seed is protected and preserved without exposing the seedling to harsh climatic condition. With a suitable environmental stimulus, the seed becomes competent to develop
4. Generally, dormancy can be seen as adaptation to survive under extreme stress, which a plant acquired during evolution
5. Evolutionary outcome: Prevalent over wide range of plant species and prominent in plants which are far away from the equator. This phenomenon is seen as an adaptation pertaining to evolution to ensure the development and differentiation of different species of plants.
6. A resting state: This state of inactivation is just a wait for a favorable condition and the seed does not lose its capacity and maintains a minimal metabolic activity as that takes place in non – dormant seeds.
Types of Seed Dormancy
Generally, Dormancy can be classified based on the type of inhibition;
1. Eco Seed Dormancy: The inhibition to growth is provided by the absence or minimal exposure of any external factors.
For Example: under cold temperature the mature onion bulbs enter a resting state.
2. Para Seed Dormancy: The growth is inhibited by other parts of the same plant.
For Example: Apical dominance of the apical bud to suppress the lateral bud development.
3. Endo Seed Dormancy: The inhibition is present in the plant part. The seed or bud or the tuber even on optimal condition fails to germinate.
The dormancy is caused by For Example: Seed coat becomes unbreakable or the embryo is efficient to the proper stimuli.
Endo dormancy further divided as Developmental Endo dormancy or Seasonal Endo Dormancy; where the dormancy is caused by the immaturity of the embryo and by intrinsic response to the climatic condition of the environment respectively.
Seed dormancy can be imposed in stages: Primary and Secondary
a. Primary Seed Dormancy: Dormancy expressed by a mature, newly dispersed seed due to endogenous inhibition like Hard coat induced dormancy or Immature embryo dormancy. The seed remains dormant even under favorable condition
b. Secondary Seed Dormancy: Dormancy is forced due to unfavorable exposure of environmental condition after seed germinates inhibiting its development.
a. Primary Seed Dormancy
(i) Coat Related Seed Dormacy
Coat related seed dormacy or Coat enhanced dormancy arises due to hard seed coat. This type is prevalent in families Fabaceae and Malvaceae.
Hard seed coat imposes various physiological and mechanical restraints for a seed to germinate. Hard seed coat is a defensive function to protect the embryo of many plants.
It also constitutes to wear and tear during dispersal mechanism. The defense functions are species specific.
Different species develops different seed coat to protect them from the harsh conditions.
For example: Xanthium – seeds are impermeable for gases, certain seeds are coated with waxes to prevent water imbibition, Conifers develops megagametophyte which prevent entry of water.
Mesquite – has lignified Testa, if a seed coat lacks Testa the endospermal wall becomes rich in hemicellulose. These are certain plants with different mechanism to protect the seed.
Such hard protecting coat can be impenetrable for the radicle causing a constraint to germinate.
Such hard coat will prevent water imbibition, gaseous exchange, retains the inhibitory components such as ABA (abscisic acid).
Identifying the reason behind dormancy will help to break the dormancy in vitro. But naturally the seeds have natural dormancy breakers in the environment.
It also depends upon the dispersal mechanism. Certain seed may require animal gut digesting enzymes to break open the seed coat.
Other seeds have cellular enzymes present in the soil aids in seed coat opening. Microorganisms in soil or in any medium where the seed develops are responsible for coat disintegration.
Inhibitions can be classified as;
1. Mechanical restraint
2. Gaseous exchange intervention
3. Retention of inhibitors
1. Mechanical restraint
Seeds with hard seed coat impose a mechanical constraint for the radicle to develop. This is broken by cellular digesting enzymes present in the soil.
For Example: Coffee seeds.
2. Gaseous exchange intervention
Phenolic compounds present in certain seeds take up high amount of oxygen content. The developing embryo receive low oxygen because of the phenolic compounds requires a high amount of oxygen to develop into plant.
Exposing such seeds to high oxygen environment will induce germination.
3. Retention of inhibitors
To prevent pre mature development of radicle, ABA is present in the seed coat inhibiting germination. These inhibitors are secondary metabolites.
Apart from ABA, Phenolic Acid, Coumarin and tannins are also additional inhibitors present at different species. these inhibitors prohibit germination.
Overcoming such condition through continuous rinsing of seeds in water will induce germination, provided optimal growth factors.
Seed Dormancy due to hard seed coat can be germinated by removing seed coat mechanically or chemically. A method which is employed under a favorable condition to remove the seed coat and induce germination is called as Scarification.
Mechanically, the coat can be removed by scratching it with knife or shaking against a coarse surface.
Chemically, a strong acid (i.e.) Sulphuric acid or organic solvent such as alcohol or acetone can be used to remove the seed coat inhibiting growth.
In few cases even boiling the seed in water eliminates the seed coat and initiate germination.
(ii) Immature Embryo Related Seed Dormacy
When a seed dehisces from the plant, there are two cases in which a seed might be present.
Case 1: the seed is matured and then released.
Case 2: Seed is still immature during dispersal.
Seeds with immature embryo belongs to Fraxinus and Anemone comes under case 2. Immaturity causes dormancy in seeds.
Under unfavourable condition they seeds might decay. the seeds reach maturation by a process of stratification. the maturation is termed as after – ripening.
Stratification takes place during autumn where the seeds are stored in layers of autumn soil, debris and snow. the cold and moisture will mature the seeds and allow them to germinate.
Artificially, the seeds are kept in the layers of sand and sphagnum which will provide them necessary conditions to germinate.
The process of after – ripening is species specific and along with stratification the seeds mature. Dormancy in certain other plants is broken by dry storage.
By the hormone balance theory, the seed has a network of hormones regulating germination.
Abscisic Acid, Gibberellins, Brassanosteroids and Ethylene form an integrated network in regulating the germination and dormancy.
Dormancy of the seed is promoted by ABA. ABA is produced from the maternal tissue to regulate the dormancy by preventing precocious germination.
For a seed to germinate the ratio of GA and ABA must be maitained.GA must be higher than ABA to over throw the inhibitory effects of ABA.
ABA also inhibits ethylene but presence of Brassanosteroids promotes ethylene which is essential for germination.
a. Secondary Seed Dormancy
Secondary dormancy is prevalent when the seed is physiologically fit and environmental factors governing the germination are absent causing a stress in seed.
Factors Causing Secondary Seed Dormancy
1. Light: Light is not an essential source supporting the germination, as the process takes place underground. But absence of light in certain Lettuce causes dormancy in seeds.
These are Positive Photoblastic seeds where the presence of light induces germination. Positive Photoblastic seeds require 660 mµ provided by red light.
Far red light at 735mµ inhibits the germination. Photo regulation is governed by pigment phytochrome which absorbs the light and detects the changes.
Similarly, germination of certain seeds takes place in dark environment and are called as negative photoblastic seeds.
2. Water: Water imbibition in seeds is a prerequisite for germination. Seeds which lack imbibition fails to germinate.
3. High CO2: High CO2 inhibits the germination as O2 is essential for regular metabolic activities of the seeds, excess CO2 inhibits the uptake of O2 causing dormancy.
4. Temperature: Temperature requirement varies for each plant. Certain plants require higher temperature for germination and others require moderate temperature.
Plants are also sensitized for low temperature treatment as in stratification before they are sown in land. Unavailability of such factors induce a secondary dormancy in mature embryo bearing seeds.
Seed Dormancy Citations
- The roles of auxin in seed dormancy and germination. Yi Chuan . 2016 Apr;38(4):314-22.
- What kind of seed dormancy occurs in the legume genus Cassia? Sci Rep . 2020 Jul 22;10(1):12194.
- Primary seed dormancy: a temporally multilayered riddle waiting to be unlocked. J Exp Bot . 2017 Feb 1;68(4):857-869.
- The Long-Standing Paradox of Seed Dormancy Unfolded? Trends Plant Sci . 2019 Nov;24(11):989-998.
- The role of light in regulating seed dormancy and germination. J Integr Plant Biol . 2020 Sep;62(9):1310-1326.
- Seed dormancy and germination. Curr Biol . 2017 Sep 11;27(17):R874-R878.
Share
Similar Post:
-

What is Fungi? Definition, Classification, Structure, and...
Continue ReadingWhat are Fungi?
Fungus (pl. fungi) is a Latin word which means mushrooms. Fungi are nucleated, spore bearing, achlorophyllous life forms which for the most part duplicate physically and abiogenetically, and whose generally filamentous stretched substantial constructions are regularly encircled by cell dividers containing cellulose or chitin, or both (Alexopoulos, 1952).
In more straightforward words it might likewise be characterized as “non-green, nucleated thallophytes”.
The normal instances of fungi are the yeasts, molds, mushrooms, polypore’s, puff balls, rusts and mucks.
The part of herbal science that arrangements with the investigation of fungi is known as mycology (Gr. mykes = mushroom + logos = talk) and the individual realizing fungi is known as mycologist.
The Italian botanist Pier’ Antonio Micheli merits the honor of being called ‘Organizer of the study of mycology’ since he was the primary individual to give substantial portrayal of fungi in his book Nova plant-arum Genera distributed in 1729.
Anton De Bary (1831-1888) is known as the ‘father of present day mycology’.
At present around 5100 genera and in excess of 50,000 types of fungi are known.
Characterization of Fungi
As per the proposals of the council on international principles of Botanical Nomenclature;
(a) It should end in—mycota.
(b) The name of regions should end in—mycotina.
(c) The name of classes should end in—mycetes.
(d) The name of subclasses should end in—mycetideae.
(e) The name of requests should end in—beers.
(f) The name of families should end in an addition—aceae.
Genera and species have no standard endings. The name of an organic entity is binomial.
It is created to parts―the first is thing assigning the family in which the organic entity has been grouped, and the second is regularly a modifier portraying the thing which signifies the species.
The primary letter of every nonexclusive name is consistently a capital.
Fungi Grouping Proposed by Linnaeus
Linnaeus (1753) in his Species Plantarum isolated the plant realm into 25 classes, which incorporate a class Crytogamia managing all plants with covered regenerative organs.
Cryptogams were additionally isolated into thallophyta, bryophyte and pteridophyta by Eichler (1886). He further partitioned thallophyta into green growth and fungi. The fungi contained Schizomycetes, Eumycetes and Lichens.
Morphology of Fungi
Monochrome micrograph showing Penicillium hyphae as long, straightforward, tube-like constructions a couple of micrometers across.
Conidiophores branch out along the side from the hyphae, ending in heaps of phialides on which circular condidiophores are orchestrated like dabs on a string.
Septa are faintly apparent as dull lines crossing the hyphae.
Most fungi develop as hyphae, which are tube shaped, string like constructions 2–10 µm in width and up to a few centimeters long.
Hyphae develop at their tips (apices); new hyphae are regularly shaped by rise of new tips along existing hyphae by an interaction called spreading, or every so often developing hyphal tips fork, bringing about two equal developing hyphae.
Hyphae likewise now and then breaker when they come into contact, a cycle called hyphal combination (or anastomosis). These development measures lead to the improvement of a mycelium, an interconnected organization of hyphae. Hyphae can be either septate or coenocytic.
Septate hyphae are partitioned into compartments isolated by cross dividers (interior cell dividers, called septa, that are framed at right points to the cell divider giving the hypha its shape), with every compartment containing at least one cores; coenocytic hyphae are not compartmentalized.
Septa have pores that permit cytoplasm, organelles, and some of the time cores to elapse through; a model is the dolipore septum in fungi of the phylum Basidiomycota.
Coenocytic hyphae are generally multinucleate supercells.
Numerous species have created particular hyphal structures for supplement take-up from living hosts; models remember haustoria for plant-parasitic types of most contagious phyla, and arbuscules of a few mycorrhizal fungi, which infiltrate into the host cells to burn-through nutrients.
"The fungi contained Schizomycetes, Eumycetes and Lichens"
Fungal mycelia can become noticeable to the unaided eye, for instance, on different surfaces and substrates, like moist dividers and ruined food, where they are generally called molds.
Mycelia developed on strong agar media in lab petri dishes are typically alluded to as provinces. These provinces can show development shapes and tones (because of spores or pigmentation) that can be utilized as symptomatic highlights in the recognizable proof of species or gatherings.
Some individual contagious provinces can arrive at unprecedented measurements and ages as on account of a clonal state of Armillaria solidipes, which reaches out over a space of in excess of 900 ha (3.5 square miles), with an expected time of almost 9,000 years.
The apothecium—a specific design significant in sexual generation in the ascomycetes—is a cup-molded organic product body that is frequently naturally visible and holds the hymenium, a layer of tissue containing the spore-bearing cells.
The basidiomycetes (basidiocarps) and a few ascomycetes are notable as mushrooms.
"The normal instances of fungi are the yeasts, molds, mushrooms, polypore's, puff balls, rusts and mucks"
Fungal generation is perplexing, mirroring the distinctions in ways of life and hereditary cosmetics inside this assorted realm of life forms.
It is assessed that 33% of all fungi recreate utilizing more than one technique for spread; for instance, propagation may happen in two very much separated stages inside the existence pattern of an animal types, the teleomorph and the anamorph.
Natural conditions trigger hereditarily resolved formative expresses that lead to the making of particular designs for sexual or asexual multiplication.
These constructions help multiplication by effectively scattering spores or spore-containing propagules.
Other than customary sexual proliferation with meiosis, certain fungi, like those in the genera Penicillium and Aspergillus, may trade hereditary material by means of parasexual measures, started by anastomosis among hyphae and plasmogamy of parasitic cells.
The recurrence and relative significance of parasexual occasions is indistinct and might be lower than other sexual cycles.
It is known to assume a part in intraspecific hybridization and is possibly needed for hybridization between species, which has been related with significant occasions in fungal advancement.
Fungi Citations
- Breakpoint: Cell Wall and Glycoproteins and their Crucial Role in the Phytopathogenic Fungi Infection. Curr Protein Pept Sci . 2020;21(3):227-244.
- Advances in Genomics of Entomopathogenic Fungi. Adv Genet . 2016;94:67-105.
- Fungi in Deep Subsurface Environments. Adv Appl Microbiol . 2018;102:83-116.
- Nematode-Trapping Fungi. Microbiol Spectr . 2017 Jan;5(1).
- Fungi in the healthy human gastrointestinal tract. Virulence . 2017 Apr 3;8(3):352-358.
- Fungi that Infect Humans. Microbiol Spectr . 2017 Jun;5(3).
Share
Similar Post: