Category: Biology
-

Epstein Barr Virus (EBV): Symptoms, Prevention, and...
Continue ReadingEpstein Barr Virus (EBV)
The Epstein Barr Virus (EBV), additionally called Human herpesvirus 4 (HHV-4), is a virus of the herpes family (which incorporates Herpes simplex virus and Cytomegalovirus) and is perhaps the most widely recognized viruses in people.
The vast majority become tainted with EBV, which is regularly asymptomatic however ordinarily causes irresistible mononucleosis.
EBV is named after Michael Epstein and Yvonne Barr, who along with Bert Achong, found the virus in 1964.
Structure of Epstein-Barr Virus (EBV)
Like other herpesviruses, Epstein Barr virus (EBV) has a toroid-formed protein center that is wrapped with DNA, a nucleocapsid with 162 capsomers, a protein covering between the nucleocapsid and the envelope and an external envelope with outer glycoprotein spikes.
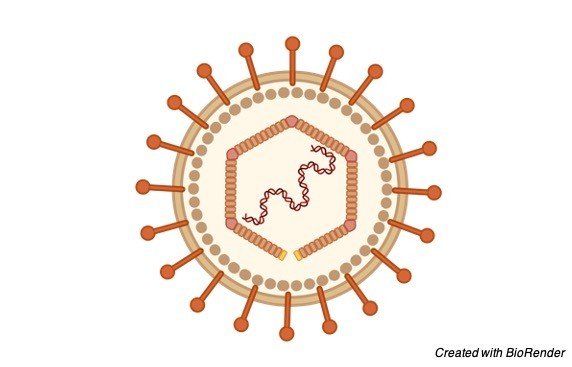
The significant EBV capsid proteins are 160, 47 and 28 kDa, comparable in size to the significant capsid proteins of herpes simplex virus. The most plentiful EBV envelope and covering proteins are 350/220 and 152 kDa, separately.
Epstein Barr Virus (EBV) Infection
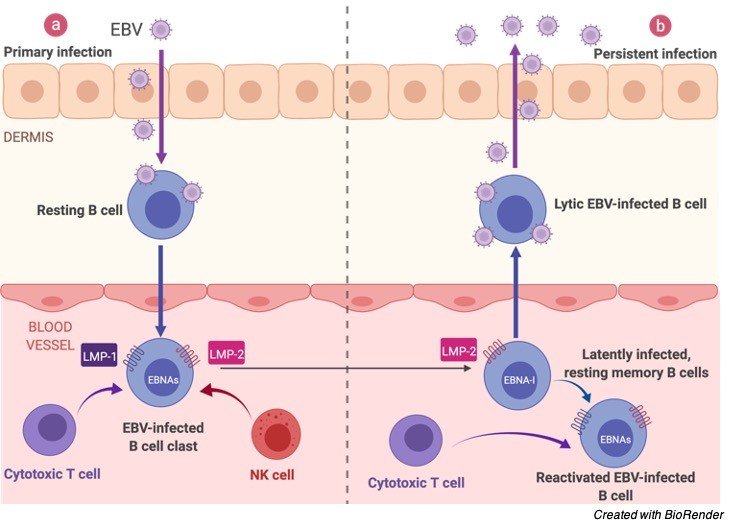
On contaminating the B-lymphocyte, the straight virus genome circularizes, and the virus therefore perseveres inside the cell as an episome.
The virus can execute numerous unmistakable projects of gene articulation which can be comprehensively classified as being lytic cycle or idle cycle.
The lytic cycle or useful disease brings about arranged articulation of a large group of viral proteins with a definitive goal of creating irresistible virions. Officially, this period of disease doesn’t unavoidably prompt lysis of the host cell as EBV virions are created by growing from the tainted cell.
The inactive cycle (lysogenic) programs are those that don’t bring about creation of virions. An exceptionally restricted, unmistakable arrangement of viral proteins are delivered during inactive cycle contamination.
These incorporate Epstein-Barr atomic antigen (EBNA)- 1, EBNA-2, EBNA-3A, EBNA-3B, EBNA-3C, EBNA-pioneer protein (EBNA-LP) and dormant layer proteins (LMP)- 1, LMP-2A and LMP-2B and the Epstein-Barr encoded RNAs (EBERs).
Also, EBV codes for no less than twenty microRNAs which are communicated in inactively contaminated cells.
Epstein Barr Virus (EBV) Gene Structure
From investigations of EBV gene articulation in refined Burkitt’s lymphoma cell lines, no less than three projects exist:
EBNA1 just (bunch I)
EBNA1 + EBNA2 (bunch II)
Dormant cycle proteins (bunch III).
It is additionally hypothesized that a program in which all popular protein articulation is stopped exists. At the point when EBV contaminates B-lymphocytes in vitro, lymphoblastoid cell lines in the long run arise that are equipped for endless development.
The development change of these cell lines is the outcome of viral protein articulation. EBNA-2, EBNA-3C and LMP-1 are fundamental for ‘change’ while EBNA-LP and the EBERs are not.
The EBNA-1 protein is fundamental for support of the virus genome. It is proposed that after normal contamination with EBV, the virus executes a few or the entirety of its collection of gene articulation projects to build up a diligent disease.
Given the underlying shortfall of host invulnerability, the lytic cycle creates a lot of viruses to contaminate other (probably) B-lymphocytes inside the host.
The dormant projects reinvent, and sabotage tainted B-lymphocytes to multiply and carry contaminated cells to the destinations at which the virus apparently endures.
Ultimately, when have resistance creates, the virus continues by winding down most (or conceivably the entirety) of its genes, just sometimes reactivating to deliver new virions.
An equilibrium is ultimately struck between intermittent viral reactivation and host insusceptible observation eliminating cells that actuate viral gene articulation.
Epstein Barr Virus (EBV) Infection Cycle
The site of industriousness of EBV might be bone marrow. EBV-positive patients who have had their own bone marrow supplanted with bone marrow from an EBV-negative giver are discovered to be EBV-negative after transplantation.
Epstein Barr Virus (EBV) Inactive Antigens
All EBV proteins are created by elective joining of a record beginning at either the Cp or Wp advertisers at the left finish of the genome (in the regular terminology).
The genes are requested EBNA-LP/EBNA-2/EBNA-3A/EBNA-3B/EBNA-3C/EBNA-1 inside the genome.
The inception codon of the EBNA-LP coding area is made by a substitute join of the atomic protein record. Without this commencement codon, EBNA-2/EBNA-3A/EBNA-3B/EBNA-3C/EBNA-1 will be communicated relying upon which of these genes is on the other hand grafted into the record.
EBNA-1
EBNA-1 protein ties to a replication beginning (oriP) inside the viral genome and intercedes replication and parceling of the episome during division of the host cell.
It is the solitary viral protein communicated during bunch I idleness. EBNA-1 has a glycine-alanine rehash that hinders antigen handling and MHC class I-confined antigen show in this manner restraining the CD8-limited cytotoxic T-cell reaction against virus tainted cells.
EBNA-1 was at first recognized as the objective antigen of sera from rheumatoid joint inflammation patients (rheumatoid joint pain related atomic antigen; RANA).
EBNA-2
EBNA-2 is the fundamental viral transactivator, changing record from the Wp advertisers utilized during at first after contamination to the Cp advertiser.
Along with EBNA-3C, it likewise initiates the LMP-1 advertiser. It is known to tie the host RBP-Jκ protein that is a central member in the Notch pathway. EBNA-2 is fundamental for EBV-intervened development change.
EBNA-3A/EBNA-3B/EBNA-3C
These genes additionally tie the host RBP-Jκ protein.
EBNA-3C
EBNA-3C is additionally a ubiquitin-ligase and has been displayed to target cell cycle controllers like pRb.
LMP-1
LMP-1 is a six-length transmembrane protein that is additionally fundamental for EBV-intervened development change. LMP-1 intervenes motioning through the Tumor rot factor-alpha/CD40 pathway.
LMP-2A/LMP-2B
LMP-2A/LMP-2B are transmembrane proteins that demonstration to obstruct tyrosine kinase flagging. It is accepted that they act to repress enactment of the viral lytic cycle.
It’s obscure whether LMP-2B is needed for EBV-intervened development change, while various gatherings have detailed that LMP-2A then again is, or alternately isn’t required for change.
EBER-1/EBER-2
EBER-1/EBER-2 are little RNAs of an obscure job. They are not needed for EBV-intervened development change.
miRNAs
EBV microRNAs are encoded by two records, one set in the BART gene and one set close to the BHRF1 bunch. The three BHRF1 miRNAS are communicated during type III dormancy while the enormous group of BART miRNAs (up to 20 miRNAs) are communicated during type II inertness. The elements of these miRNAs are at present obscure.
Epstein Barr Virus (EBV) Surface Receptors
The Epstein-Barr Virus surface glycoprotein H (gH) is fundamental for infiltration of B cells yet in addition assumes a part in connection of virus to epithelial cells.
In lab and creature preliminaries in 2000, it was shown that both opposition of RA-intervened development restraint and advancement of LCL expansion were productively switched by the glucocorticoid receptor (GR) bad guy RU486.
Sicknesses Related With Epstein Barr Virus (EBV)
• Irresistible states
• Irresistible mononucleosis
• Stevens-Johnson condition
• Hepatitis
• Herpes
• Histiocytic necrotizing lymphadenitis (otherwise called Kikuchi’s infection)
Diseases Caused by Epstein Barr Virus (EBV)
• Non-Hodgkin’s lymphomas, including essential cerebral lymphoma
• Hodgkin’s infection • hairy leukoplakia
• Nasopharyngeal carcinoma
• Smooth muscle tumors
• Immunocompromised/stifled states: Normal variable immunodeficiency
Summary Epstein Barr Virus (EBV)
Much of the time of irresistible mononucleosis, the clinical conclusion can be produced using the trademark group of three of fever, pharyngitis, and lymphadenopathy going on for 1 to about a month.
Serologic test outcomes incorporate an ordinary to reasonably raised white platelet tally, an expanded complete number of lymphocytes, more prominent than 10% abnormal lymphocytes, and a positive response to a “mono spot” test.
In patients with manifestations viable with irresistible mononucleosis, a positive Paul-Bunnell heterophile immunizer test result is demonstrative, and no further testing is essential.
Moderate-to-undeniable degrees of heterophile antibodies are seen during the principal month of ailment and decline quickly after week.
Bogus positive outcomes might be found in few patients, and bogus adverse outcomes might be acquired in 10% to 15% of patients, fundamentally in youngsters more youthful than 10 years old.
Genuine episodes of irresistible mononucleosis are amazingly uncommon. At the point when “mono spot” or heterophile test results are negative, extra research center testing might be expected to separate EBV contaminations from a mononucleosis-like sickness prompted by cytomegalovirus, adenovirus, or Toxoplasma gondii.
Direct recognition of EBV in blood or lymphoid tissues is an examination instrument and is not accessible for routine determination.
All things considered; serologic testing is the technique for decision for diagnosing disease.
Epstein Barr Virus (EBV) Citations
- Epstein Barr virus strains and variations: Geographic or disease-specific variants? J Med Virol . 2017 Mar;89(3):373-387.
- Epstein Barr virus (EBV) status in colorectal cancer: a mini review. Hum Vaccin Immunother . 2019;15(3):603-610.
- Epstein Barr virus strain variation and cancer. Cancer Sci . 2019 Apr;110(4):1132-1139.
- Epstein Barr Virus. Microbiol Spectr . 2016 Jun;4(3).
- Epstein Barr virus (EBV) reactivation and therapeutic inhibitors. J Clin Pathol . 2019 Oct;72(10):651-658.
Share
Similar Post:
-

Gram-Negative Bacteria: Definition, Structure, and Facts
Continue ReadingWhat are Gram-Negative Bacteria?
Gram-negative bacteria don’t hold the violet stain utilized within the gram-staining technique for bacterial differentiation.
They are portrayed by their cell envelopes, which are made out of a flimsy peptidoglycan cell divider sandwiched between an inward cytoplasmic cell membrane and a bacterial external membrane.
Gram-negative bacteria are found in basically all conditions on Earth that help life.
The gram-negative bacteria incorporate the model creature Escherichia coli, just as numerous pathogenic bacteria, for example, Pseudomonas aeruginosa, Chlamydia trachomatis, and Yersinia pestis.
They are a significant clinical test, as their external membrane shields them from numerous anti-infection agents (counting penicillin); cleansers that would typically harm the peptidoglycans of the (internal) cell membrane; and lysozyme, an antimicrobial compound delivered by creatures that structures part of the natural safe framework.
Also, the external handout of this membrane involves a complex lipopolysaccharide (LPS) whose lipid A segment can cause a poisonous response when these bacteria are lysed by invulnerable cells.
This poisonous response can incorporate fever, an expanded respiratory rate, and low pulse—a perilous condition known as septic shock.
A few classes of anti-microbials have been intended to target gram-negative bacteria, including aminopenicillins, ureidopenicillins, cephalosporins, beta-lactam-betalactamase inhibitor blends (for example piperacillin-tazobactam), Folate bad guys, quinolones, and carbapenems.
Large numbers of these anti-infection agents additionally cover gram-positive creatures. The medications that explicitly target gram negative life forms incorporate aminoglycosides, monobactams (aztreonam) and ciprofloxacin.
Gram-Negative Bacteria Cell Structure
Gram-negative bacteria are portrayed by the presence of the periplasmic space, which is a solitary layer of peptidoglycan sandwiched between the cytoplasmic membrane and the external membrane.
Peptidoglycan, otherwise called murein, is a polymer that comprises of a carbohydrate structure and amino acids.
Peptide chains inside the peptidoglycan structure are halfway cross-connected in Gram-negative bacteria, diverging from the profoundly cross-connected peptide chains of Gram-positive bacteria.
The external membrane contains lipopolysaccharide, an enormous molecule that is poisonous to creatures.
During Gram staining, the external membrane of Gram-negative bacteria weakens from the liquor added to the example and the slight layer of peptidoglycan can’t hold the gem violet stain.
The counterstain is added to give contrast by staining the decolorized Gram-negative bacteria through the dainty peptidoglycan layer while being adequately light to not upset the precious stone violet staining on the Gram positive bacteria.
Etiology of Gram-Negative Bacteria
The gram-negative bacteria have an incredible capacity to cause sickness in people and can arrive at practically all frameworks in the living being, like the stomach related framework, sensory system, urinary framework, and circulation system, causing diarrheal gastroenteritis until extreme meningitis.
Such microorganisms colonize the digestion tracts, aviation routes, and skin, which favors the spread to different pieces of the human organic entity, particularly in immunocompromised people.
Probably the best trouble of wellbeing experts is to treat nosocomial contaminations of the lower respiratory parcel in which the microbes GNB are included on the grounds that despite the fact that they are liable for a decent bit of these diseases, they are non-receptive to anti-infection treatment because of the great opposition rates and the helpless infiltration of medications into the lung parenchyma.
Another significant concern is gastroenteritis brought about by Enterobacteriaceae (Shigella spp., Salmonella spp., enteropathogenic E. coli), which influences a huge number of individuals worldwide and is identified with absence of disinfection.
Also, they are liable for meningitis – a conceivably lethal infection if not treated on schedule – it obtained both locally and in the medical clinic climate.
Urinary lot diseases are likewise normal, particularly in young ladies. In any case, these diseases turned into an issue with the wild development of multi-safe bacteria.
At last, bacteremia is a significant intricacy of these diseases, likewise due to the previously mentioned opposition that the microorganisms illustrated.
Assessment of Gram-Negative Bacteria
While the contaminations delivered by gram-negative are basically undefined from different etiologies, the lab portrayal can characterize the organic entity and its antimicrobial weakness profile.
In this manner, the highest quality level assessment of these diseases are the way of life; notwithstanding, they have the issue of a deferral in the outcome.
Another fundamental test for bacterial separates is the Gram stain; albeit straightforward, it can rapidly recognize the course of a medication mediation.
Enterobacteria, on account of their heterogeneity, rely upon different biochemical tests (Indol, Voges-Proskauer, catalase, cytochrome oxidase, urease, motility, citrate, ONPG, Dnase, decarboxylation of lysine, gas creation, among others), filled in essential detachment agar.
Species like E. coli, in light of the fact that they are fermenters of solid acids, they produce enormous amounts of blended acids, which make the pH of the media present pink states.
Others, like non-fermenters, fill in essential detachment media (blood agar and MacConkey), in the OF-oxidation/aging medium – fill just in the high-impact climate.
Be that as it may, notice the recognizable proof of bacterial strains impervious to numerous medications may require sub-atomic techniques, however they are not accessible in all research centers.
Phenotypic strategies like Modified Hodge (MHT) and Combined Diffusion Disk (CDT) utilizing EDTA are a decent other option.
The MHT is a test dependent on the inactivation of carbapenems by bacterial strains containing the catalysts carbapenemases, empowering a vulnerable strain to stretch out its development to a circle containing the antimicrobial along the inoculum of the strain tried.
This test is suggested for strains with high least inhibitory fixation (MIC) or diminished zones of hindrance in the plate.
The CDT includes a test plate dispersion with carbapenem anti-microbials, where carbapenem circles are put with and without EDTA, and their association predicts whether the organic entity creates or doesn’t carbapenemase.
Treatment/Management of Gram-Negative Bacteria
Treatment choices for gram-negative MDR contaminations are restricted, and the outcomes are by and large baffling a result of medication obstruction.
MDRs are as yet advancing, which gives protection from novel antimicrobials. Accordingly, as new medications’ accessibility advances gradually, some recently deserted alternatives have returned, like polymyxins and colistin that have high poisonousness (nephrotoxicity and neurotoxicity).
Moreover, safe qualities to these medications have been accounted for, for example, mcr-1 – cause extra concern.
However, it is realized that the mix of these medications with carbapenems may have a worked on synergistic activity. Another medication of decision is tigecycline since it shows in vitro movement against MDR; notwithstanding, there are restrictions in its utilization as high portions are required, and tissue entrance is regularly poor, which impedes its activity in vivo.
Gram-Negative Bacteria Citations
- Multidrug-resistant Gram-negative bacteria: a product of globalization. J Hosp Infect . 2015 Apr;89(4):241-7.
- Potent Antibiotics Active against Multidrug-Resistant Gram-Negative Bacteria. Chem Pharm Bull (Tokyo) . 2020;68(3):182-190.
- Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol . 2014;9(10):1165-77.
- Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: a review. J Chemother . 2017 Dec;29(sup1):2-9.
- Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol . 2016 Aug 11;14(9):576-88.
Share
Similar Post:
-

Gram-Positive Bacteria: Definition, Structure, and Facts
Continue ReadingWhat is Gram-Positive Bacteria?
In bacteriology, gram-positive bacteria produces a positive outcome in the Gram stain test, which is generally used to rapidly arrange bacteria into two general classifications as per their sort of cell.
Gram-positive bacteria take up the violet stain utilized in the test, and afterward have all the earmarks of being purple-shaded when seen through an optical magnifying instrument.
This is on the grounds that the thick peptidoglycan layer in the bacterial cell divider holds the stain after it is washed away from the remainder of the example, in the decolorization phase of the test.
On the other hand, gram-negative bacteria can’t hold the violet stain after the decolorization step; liquor utilized in this stage debases the external layer of gram-negative cells, making the cell divider more permeable and unequipped for holding the precious stone violet stain.
Their peptidoglycan layer is a lot slenderer and more sandwiched between an internal cell film and a bacterial external layer, making them take up the counterstain (safranin or fuchsine) and seem red or pink.
Notwithstanding their thicker peptidoglycan layer, gram-positive bacteria are more responsive to certain cell wall focusing on anti-toxins than gram-negative bacteria, because of the shortfall of the membrane present outside.
Etiology of Gram-Positive Bacteria
Gram-positive cocci incorporate Staphylococcus (catalase-positive), which develops bunches, and Streptococcus (catalase-negative), which fills in chains.
The staphylococci further partition into coagulase-positive (S. aureus) and coagulase-negative (S. epidermidis and S. saprophyticus) species. Streptococcus bacteria partition into Strep. pyogenes (Group A), Strep. agalactiae (Group B). Gram-positive bacilli (rods) partition as per their capacity to deliver spores.
Bacillus and Clostridia are spore-framing rods while Listeria and Corynebacterium are not. Spore-framing rods that produce spores can make due in conditions for a long time.
Additionally, the stretching fiber rods envelop Nocardia and actinomyces. Gram-positive living beings have a thicker peptidoglycan cell wall contrasted and gram-negative bacteria.
It is a 20 to 80 nm thick polymer while the peptidoglycan layer of the gram-negative cell wall is 2 to 3 nm thick and covered with an external lipid bilayer film.
Circulatory system disease death rates have expanded by 78% in only twenty years. Gram-positive life forms have exceptionally factor development and opposition designs.
The SCOPE project (Surveillance and Control of Pathogens of Epidemiologic Importance) found that gram-positive living beings in those with a fundamental threat represented 62% of all circulatory system diseases in 1995 and 76% in 2000 while gram-negative life forms represented 22% and 14% of contaminations for these years.
Pathophysiology of Gram-Positive Bacteria
Staphylococcus aureus is a gram-positive, catalase-positive, coagulase-positive cocci in groups. S. aureus can cause incendiary sicknesses, including skin contaminations, pneumonia, endocarditis, septic joint pain, osteomyelitis, and abscesses.
S. aureus can likewise cause poisonous shock disorder (TSST-1), singed skin condition (exfoliative poison, and food contamination (enterotoxin).
Staphylococcus epidermidis is a gram-positive, catalase-positive, coagulase-negative cocci in bunches and is novobiocin touchy.
S. epidermidis ordinarily contaminates prosthetic gadgets and IV catheters creating biofilms. Staphylococcus saprophyticus is novobiocin safe and is a typical verdure of the genital plot and perineum. S. saprophyticus represents the second most normal reason for simple urinary lot contamination (UTI).
Streptococcus pneumoniae is a gram-positive, epitomized, lancet-formed diplococci, most generally causing otitis media, pneumonia, sinusitis, and meningitis. Streptococcus viridans comprise of Strep. mutans and Strep mitis found in the ordinary vegetation of the oropharynx ordinarily cause dental conveys and subacute bacterial endocarditis (Strep. sanguinis).
Streptococcus pyogenes is a gram-positive gathering A cocci that can cause pyogenic diseases (pharyngitis, cellulitis, impetigo, erysipelas), toxigenic contaminations (red fever, necrotizing fasciitis), and immunologic diseases (glomerulonephritis and rheumatic fever). ASO titer distinguishes S. pyogenes diseases.
Streptococcus agalactiae is a gram-positive gathering B cocci that colonize the vagina and is discovered chiefly in infants.
Pregnant ladies need evaluating for Group-B Strep (GBS) at 35 to 37 weeks of incubation. Enterococci is a gram-positive gathering D cocci discovered essentially in the colonic vegetation and can cause biliary lot contaminations and UTIs. Vancomycin-safe enterococci (VRE) are a significant reason for nosocomial diseases.
Gram-Positive Bacteria Rod
Clostridia is a gram-positive spore-shaping bar comprising of C. tetani, C. botulinum, C. perfringens, and C. difficile. C. difficile is frequently optional to anti-microbial use (clindamycin/ampicillin), PPI use, and late hospitalization.
Treatment includes principally with oral vancomycin. Bacillus anthracis is a gram-positive spore-framing bar that produces Bacillus anthracis poison bringing about a ulcer with a dark eschar.
Bacillus cereus is a gram-positive bar that can be procured from spores making due half-cooked or warmed rice.
Indications incorporate sickness, heaving, and watery non-wicked loose bowels. Corynebacterium diphtheria is a gram-positive club-molded bar that can cause pseudomembranous pharyngitis, myocarditis, and arrhythmias. Pathogen immunizations forestall diphtheria.
Listeria monocytogenes is a gram-positive bar gained by the ingestion of cold store meats and unpasteurized dairy items or by vaginal transmission during birth.
Listeria can give rise to neonatal meningitis, gastroenteritis, and septicemia. Treatment incorporates ampicillin.
Gram-Positive Bacteria Cell Wall Structure
The cell wall construction of Gram positive bacteria comprises of the periplasmic space encased between the plasma film and a thick peptidoglycan layer.
In contrast with Gram negative bacteria, the periplasmic space of Gram positive bacteria is more modest in volume and the cell wall is a lot thicker, going from 15 and 80 nanometers.
The thick cell wall is made out of a few peptidoglycan layers, a design dependent on a glycan spine and exceptionally cross-connected peptide chains.
Another particular trait of Gram positive bacteria is the presence of teichoic acids inside the cell wall.
Teichoic acids are anionic polyol phosphate polymers that give inflexibility to the cell wall by either securing to the plasma film or through the covalent connection to peptidoglycan.
Anionic polymers additionally have comparative capacities to the external layer of Gram negative bacteria by impacting penetrability, interceding cooperations and going about as a platform for extracytoplasmic catalysts during cell-wall development.
Gram-Positive Bacteria MRSA
Gram positive bacteria are more defenseless to treatment with anti-microbials than Gram negative bacteria since they do not have an external layer, regardless certain bacterial strains show antimicrobial opposition.
Methicillin-safe Staphylococcus aureus or MRSA is a typical Gram positive microorganism that is impervious to all β-lactam antimicrobials including penicillin.
Numerous β-lactam antimicrobials are given as first line treatment for staphylococcal disease in view of their prevalent viability.
Protection from these medications implies that second line specialists are required, which confound treatment regimens and adversely sway patient results.
The impact of anti-toxins on Gram positive bacteria is decreased on account of antimicrobial opposition instruments, for example, β-lactamase creation and alterations to the objective site of the anti-toxin.
The hereditary determinants of anti-microbial opposition are communicated between bacteria through both vertical and level exchange, with a few qualities equipped for being embedded in an integron prompting the obstruction of various antimicrobials.
Restorative systems for the treatment of MRSA and other antimicrobial safe bacteria incorporate higher anti-microbial measurement, mix treatment with non-ordinary medications and the improvement of new medications.
Gram-Positive Bacteria Citations
- Gram-negative and Gram-positive bacterial extracellular vesicles. Semin Cell Dev Biol . 2015 Apr;40:97-104.
- Surface Proteins on Gram-Positive Bacteria. Microbiol Spectr . 2019 Jul;7(4):10.1128/microbiolspec.GPP3-0012-2018.
- Extracellular Vesicle Biogenesis and Functions in Gram-Positive Bacteria. Infect Immun . 2020 Nov 16;88(12):e00433-20.
- Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol . 2013;67:313-36.
Share
Similar Post:
-
Coding DNA: Definition, Function, and Structure
Continue ReadingWhat is Coding DNA?
Life’s genetic code is written in the DNA particle (also known as deoxyribonucleic acid). According to the point of view of plan, there is no human language that can coordinate with the effortlessness also, class of DNA. DNA has four significant capacities:
(1) it contains the outline for making proteins and compounds;
(2) it assumes a part in directing when the proteins and catalysts are made and when they are not made;
(3) it conveys this data when cells gap; and
(4) it sends this data from parental organic entities to their posterity. In this section, we will investigate the construction of DNA, its language, and how the outline becomes converted into an actual protein.
Coding DNA: The Genetic Code: A General Viewpoint
DNA is an outline. It doesn’t truly develop anything. Prior to examining how the data in the DNA brings about the assembling of a substantial particle, it will be it essential to acquire a general viewpoint on the genetic code.
It is helpful to see the genome for any species as a book with the genetic code as the language normal to the books of all living things.
The “letter set” for this language has four and just four letters given by four nucleotides in DNA (A,T,C, and G) or RNA (A,U,G and C). As opposed to human language, where a word is created of quite a few letters, a genetic “word” comprises of three and just three letters.
Each genetic word represents an amino corrosive. (We will characterize an amino corrosive later.) For model, the nucleotide arrangement AAG is “DNAese” for the amino corrosive phenylalanine, the arrangement GTC indicates the amino acid glutamine, and the succession AGT represents the amino acid serine.
Like normal language, DNA has equivalent words. For instance, ATA and ATG both mean the amino corrosive tyrosine.
The sentence in the DNA language is a progression of words that gives an arrangement of amino acids.
For instance, the DNA sentence AACGTATCGCAT would be perused as a polypeptide chain made out of the amino acids leucine-histidine-serine-valine. Due to the trio idea of the DNA language, it isn’t important to put spaces between the words.
Given the right beginning position, the language will interpret with 100% constancy.
Like regular composed language, part of the DNA language comprises of accentuation marks. For instance, the nucleotide DNA trios ATT, ATC, and ACT are practically equivalent to a period (.) in finishing a sentence—each of the three sign the finish of a polypeptide chain.
Other accentuation marks signify the beginning of the amino corrosive succession for the peptide. Dissimilar to the trio idea of the DNA words for amino acids, some DNA accentuation imprints may be pretty much than three nucleotides.
Coding DNA: A Complicated Design
At last, DNA, actually like a book, is coordinated into sections. The parts compare to the chromosomes, so their number will shift starting with one animal types then onto the next.
The book for people comprises of 23 unique sections or chromosomes. The book for different species may contain less or more sections with little connection between’ s the number of sections and the intricacy of the living thing.
The contrasts between regular human language and DNAese are pretty much as significant as the similitudes.
All distinctions decrease to the way that human language is sound while DNA is the most jumbled and scattered correspondence framework at any point created.
To begin with, the parts in a human language book are masterminded to recount a lucid story. There is no such requesting to chromosomes.
Second, sentences in English truly follow each other with one sentence qualifying, decorating, or adding data to another to finish a cognizant line of thought. The genetic language infrequently, if at any point, has a consistent grouping.
Figuratively, one DNA sentence may portray the climate, the following give two elements for a stew formula, and the third could be a political axiom.
Third, though it is silly to compose an English compound sentence with a section or two mixed between the two autonomous conditions, DNA regularly places free provisos of similar sentence in altogether various parts.
Fourth, no English book would be distributed where most sentences are hindered with what seems, by all accounts, to be the thoughts of a chimpanzee haphazardly striking a console.
A single DNA sentence might be punctured with over twelve long arrangements of such clear babble.
Fifth, with normal language it is viewed as awful way of talking to rehash something similar thought in adjoining sentences, let alone in similar words. With DNA redundancy is the standard, not the exemption.
At last, the size of the DNA “book” for any mammalian species far surpasses that of any book composed by a human. With eighty-a few characters for every line and thirty-a few lines on a page, a 500 page book contains around 1,500,000 English letters.
It would take more than 2,000 such books to contain the DNA book of homo sapiens. What’s more, practically 90% of the characters in these 2,000 volumes have no obvious significance!
Protein Synthesis Hidden in Coding DNA
The fundamental structure block for any protein or then again compound is the amino corrosive. There are twenty amino acids utilized in developing proteins, a large portion of which contain the addition “ine,” e.g., phenylalanine, serine, tyrosine.
Amino acids are much of the time curtailed by three letters, generally the initial three letters of the name—e.g., phe for phenylalanine, tyr for tyrosine.
There are three significant hotspots for the amino acids in our bodies. To begin with, the cells in our bodies can produce amino acids from other, more essential mixtures (or, by and large, from other amino acids).
Second, proteins and chemicals inside a cell are continually being separated into amino acids. At long last, we can get amino acids from diet.
At the point when we eat a succulent steak, the protein in the meat is separated into its amino acids by chemicals in our stomach and digestive tract.
These amino acids are then shipped by the blood to different cells in the body.
A progression of amino acids genuinely connected together is known as a polypeptide chain. Until further notice, think about a polypeptide chain as a straight series of train units coupled together.
The train units are the amino acids and their couplings the compound bonds holding them together.
The series is straight as in it doesn’t branch into a Y-like construction. The expressions polypeptide chain, polypeptide, or peptide tie typically allude to a more extended series of coupled amino acids, some of the time numbering in large numbers.
Be attentive, in any case. A protein is at least one polypeptide chains genuinely combined and taking on a three-dimensional arrangement.
The polypeptide chain(s) containing a protein will twist, overlap back upon themselves, and bond at different spots to give an atom that is as of now not a basic direct structure.
A model is a haemoglobin, a protein in the red platelets that conveys oxygen. It is made out of four polypeptide chains that twist and bond and consolidate.
Some proteins contain synthetic compounds other than amino acids. An especially significant class of proteins is the receptor protein that lives on the cell membrane and is liable for “correspondence” between the cell and extracellular “couriers.”
Coding DNA Citations
Share
Similar Post:
-

Protein Translation: Definition, Function, and Structure
Continue ReadingWhat is Protein Translation?
We all know that anything which is translated can make it easy to activate or become functionable. Like how we want us to translate the other languages into our own mother tongues so that it can be very easy for us to understand, likewise our cells also want the information to get translated in its own way to make the enzymes and the body to get activated.
Like wise protein translation is very important for the carrying out the genetic process.
Translation is a process in which protein is synthesized from the information that is being stored in the messenger RNA, during this process of translation, the mRNA sequence is being read by the genetic code, with the help of set of rules in the genomes, mRNA is translated as the 20-letter code of amino acids, which acts as a building block.
The genetic code is a set of three letter combinations of the nucleotides, which are known as codons.
Each of the codon corresponds to the specific amino acid or stop signal. Translation often occurs in a structure of called as ribosome, which is usually known as the site for the synthesis of proteins.
It is also known as protein factory. The ribosome is made up of smaller and larger sub unit and it is also a complex molecule which composed of several proteins along with the ribosomal RNAs.
Translation process in the messenger RNA occurs in three stages as initiation, elongation and termination. And they can be discussed here.
Protein Translation Features
Generally, in the molecular biology and the genetics, protein translation is the process where the ribosomes that are present in the endoplasmic reticulum and the cytoplasm synthesizes the proteins during the process of transcription of the DNA to RNA in the nucleus of the cell.
This entire process is termed as gene expression. During the translation, the messenger RNA is being decoded in a ribosome, outside a nucleus, which produces a specific amino acid chain or a polypeptide chain, which later folds into an active protein and performs its functions the ribosomes which induces the binding of complementary tRNA anticodons sequences in the mRNA by facilitating the decoding process by inducing the binding site.
The tRNAs carries a specific amino acid test which are chained together into a polypeptide as the messenger RNA by passing through and it is being read by the ribosome.
Steps of Protein Translation
Translating process takes place in 3 steps as initiation, elongation and termination.
I. Initiation
In the process of initiation, ribosome assembles itself around the target mRNA, initially tRNA attaches to the start codon.
II. Elongation
Elongation is the mid process of the translation where last tRNA which is validated by the small ribosomal sub unit helps in transferring the amino acid by carrying it to the large unit of the ribosome, which then further binds to the one of tRNA is present prior to it.
The ribosome then moves next of the mRNA codon and continues the process of translocation which creates an amino acid chain.
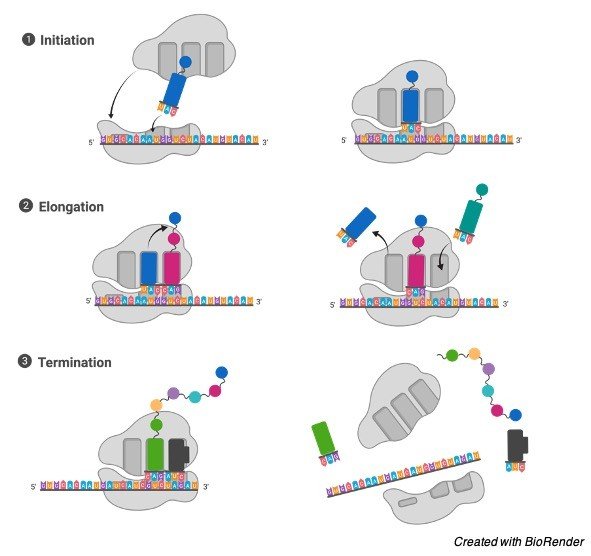
III. Termination
When a stop codon is reached finally, the ribosomes release the polypeptide. The complex of the ribosome remains intact and it furthers moves next to the mRNA to get translated.
In prokaryotes, like bacteria and archaea, the process of translation occurs in the cytosol. During this process, the large and small sub units of the ribosomes bind with the messenger RNA, where as in eukaryotes, translocation occurs in the cytoplasm and it also occurs across the membrane of the endoplasmic reticulum and this process is known as co-translational translocation.
During this trans locational process, the ribosomes binds to the outer membrane in the endo plasmic reticulum so that it is also known as rough endoplasmic reticulum.
The new protein which is synthesized is released in the endoplasmic reticulum, and the newly created polypeptide are stored within the endoplasmic reticulum for future vesicle transport and it is secreted in the cell membrane and it is used for immediate activities.
Many types of RNAs can be transcribed such as tRNA, Rrna, and the small nuclear RNA does not undergo translation to get translated into proteins.
Mechanism of Protein Translation
The basic process of protein synthesis is done by addition of one nucleotide at the same time which ends in formation of a protein.
Ribosomes play an important role in synthesizing the proteins. A ribosome is usually made up of two subunits, smaller and a larger unit, where these two units come together before a process of translation in the messenger RNA which is further translated into a protein and provides a site for the process of translation for the production of polypeptides.
The selection of the type of amino acids is decided by the messenger RNA molecule.
Where as each molecule of the added amino acid gets matched up with a three nucleotide sequences in the messenger RNA.
For the formation of each of the triplet codon, the respective amino acid should be added to the chain at match the nucleotides to form a triplet in the mRNA.
In the same manner a sequence of amino acids is made. Then further mRNA carries the information which has been encoded in the ribonucleotide sequence and they are read by the translational machinery process to form a triplet codon.
These codons then specify a specific amino acid. The messenger RNA sequence is used as the template for the assembling in an order and the chain of amino acids that form a protein.
Protein Translation Citations
- Protein Translation and Psychiatric Disorders. Neuroscientist . 2020 Feb;26(1):21-42.
- Epigenetic Regulation of the Protein Translation Machinery. EBioMedicine . 2017 Mar;17:3-4.
- Protein translation, 2008. Aging Cell . 2008 Dec;7(6):777-82.
- Protein Synthesis Initiation in Eukaryotic Cells. Cold Spring Harb Perspect Biol . 2018 Dec 3;10(12):a033092.
Share
Similar Post:
-

Codon: Definition, Function, and Mechanism
Continue ReadingWhat is Codon?
In biology, a codon refers to a basic genetic unit of the life which acts as the template for the amino acid for the production of protein expressions.
The information that are required for the is stored in the form of genes and it expresses protein for the modality through which the information is encoded on the genes to be expressed. Thus the codon is considered as an essential genetic unit of life.
A codon is a specific sequence of the nucleotides on the mRNA which corresponds to a particular amino acid.
A codon also acts as a stop signal during the process of protein translation.
We all are familiar that nucleotide is made up of a nucleobase, a sugar and a phosphate group.
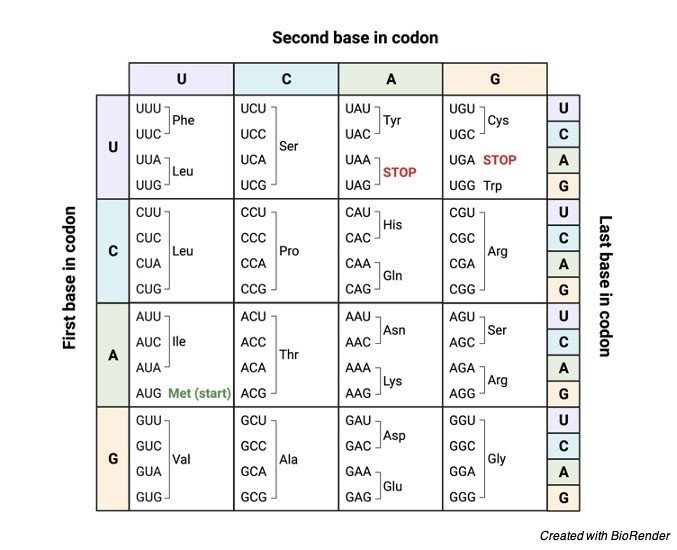
Do you all know how many of nucleotides make up the codon? Yeah, a sequence of about three nucleotides makes up a codon. In other way it can also be said that a three-nucleotide sequence make up a single codon on the mRNA.
The basic nucleotides of RNA are Adenine, Guanine, Cytosine and Uracil. You may then get into doubt how such three nucleotides are form?
A codon is typically made up of three nucleotides in a combination of AUG, UCC, UGA which all codes for a particular amino acid. And then it is also important to know what does codon codes for?
All the collection of codons on the mRNA forms a genetic code, a codon is generally a smallest unit of a genetic code, in simple terms, it can also be said that each codon encrypts a specific amino acid.
It has also had the capability of coding for a signal to start or stop the process of the protein synthesis in a particular cell.
Features of Codon
Codon is usually defined as the amino acid which acts as coding unit in the messenger RNA or in a DNA. The codons which are present in the form of strings on the mRNA describes about the order of the amino acids in an encoded protein.
Apart from these amino acids, these are also few codons which specifically works on the start and stop signals.
Each codon contains a set of three adjacent nucleotides, which are called as triplets in the base pair of the mRNA, as three bases of the corresponding anticodon of a tRNA molecule which carries a particular amino acid.
The ribosome which is considered as an important organelle for the synthesis of proteins has binding sites for the tRNAs which helps in matching their corresponding codons in the mRNA.
Types of Codon
Before understanding about the function of codon, It is very much important to know about the process of translation.
Let us consider here an example of the cellular process which explains us about the connection between a codon and amino acid.
Each of the DNA is made up of number of genes which carries the important functions that is very much essential for the life and thus generates a response for the creation of vital protein molecules.
This expression of the gene entails the synthesis of the encoded proteins through this.
Thus, the synthesis of proteins is carried out through two steps, which are known as transcription and translation.
Transcription is the first step involved in the process of synthesis of proteins. Here DNA acts as a template for generating the messenger RNA.
During this process of forming mRNA, the information that has been encoded in the DNA for each of the protein is transferred into mRNA. Thus, DNA acts as a template for a complementary base pairing for transferring the information of protein expression into the mRNA.
Translation: An mRNA is one of the single stranded nucleic acid. The genetic information that is carried in it is acquired from the molecule of DNA by the process of transcription.
The genetic code includes codons which will be translated into the proteins. Thus, the codons and the amino acids are related to each of them and are very important for processing the life.
Anticodon is also considered as a trinucleotide sequence, which are found in the tRNA.
These are nucleotide sequences that are being found complementary to the base sequence of the mRNA.
The presence of these anti codons on the transfer RNA ensures whether the suitable amnio acid is introduced into the protein structure.
It is hence cleared that the anti-codons are present on the mRNA where as the codons are present in tRNA.
Types of Codon
Generally, codons are classified into two types, as signal codons and the non-signal codons.
The signal codons are the codons which provides the signal during the process of translation, these codons are further classified as start and stop codon.
Where start codon encodes the signals foe initiating the protein translation and the stop codons helps in terminating the process of translation.
The non-signal codons are primarily used for the process of translation, which activates typically after the translating of start codons.
Codon Table
The codons can also be classified in a different way that whether the codon codes for an amino acid or not.
The codon which codes for a specific amino acid is known as sense codon and the codon which is not coding for an amino acid is known as non-sense codon.
Generally, there are about 64 codons. Out of these 64 codons, only 61 codons codes for twenty amino acids and the rest of them codes for the codon signals.
It is till surprising that 61 codons can code only for 20 amino acids and this is because the amino acids are specified particularly more than one codon. By translating the nucleotides into the amino acid.
Codon Citations
Share
Similar Post:
-
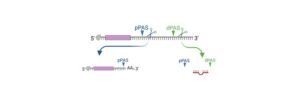
Polyadenylation: Definition, Function, and Mechanism
Continue ReadingWhat is Polyadenylation?
In most of the eukaryotes, mRNAs in the cells undergoes many complications to modify and get processed before undergoing the process of translation.
These modification process involves some chemical modifications, polyadenylation and removal of introns.
Here we will be discussing briefly about polyadenylation. Polyadenylation is a process of adding a poly(A) tail to the mRNAs, as these polyadenyl tails consists of numerous adenine monophosphates.
It is an important process in all the eukaryotes to produce a mature messenger RNA which is greatly used for transcribing. So, it forms a larger part of the gene expression process.
Poly (A) tail is very much important for the nuclear export, translation and in maintaining the stability of the mRNA.
This tail is shortened over time, and when it is shortened enough, mRNA is enzymatically degraded,
Features of Polyadenylation
Polyadenylation is a process of addition of poly(A) tail to transcript RNA, typically a messenger RNA (mRNA).
The poly (A) tail contains multiple of adenosine monophosphate, in other terms it can also be said that it is a stretch of RNA which contains only adenine bases.
In eukaryotic organisms’ polyadenylation is considered as a part of process which produces mature messenger RNA for the process of translation.
In many of the bacteria’s the poly(a) tail promotes degradation in the mRNA. Which therefore forms a part of the large processor in the gene expression.
The poly (A) tail is very much important for the nuclear export, translation and in maintain the stability of mRNA.
The tail is shortened upon time until it becomes short enough when the mRNA is enzymatically degraded. However, in some cell types mRNAs has a short poly(A) tails which are stored for the activation at the later times with the help of re-polyadenylation in the cytosol.
Where as in bacteria the polyadenylation promotes the degradation of RNA; in certain cases, this condition leads to non-coding of eukaryotic RNAs.
RNA and Polyadenylation
RNA is one of the large biomolecules, where each of them plays an important role in building the nucleotides by acting as a building block.
The term poly (A) denotes polyadenylic acid tail which reflects the RNA nucleotides for a base Adenine denoted by a letter A, and the other bases where G is noted for Guanine and C is noted for cytosine and U is noted as Uracil.
RNA is produced from the DNA during the transcribing process. During this conversion RNA sequences are written in a direction of 5’ to 3’ end.
Where 5’ end is the part of an RNA molecule which is transcribed first and later the 3’ end is transcribed.
The 3’end is the place where polyadenyle tail is found on the polyadenylated RNAs.
Messenger RNA is a kind of RNA which has a coding region and acts as a template for the synthesis of proteins which is otherwise called as translation.
Whereas the rest of the parts expect coding regions in the messenger RNAs remain as the untranslated regions and they just tunes how active the mRNA is.
This condition can be found in so many RNAs which are not being translated. And they are commonly called as non-coding RNAs.
Similar to that of the untranslated regions, many of the non-coding RNAs also performs some regulatory roles.
3' Polyadenylation
The mRNA molecules which is present in both prokaryotes and eukaryotes have the polyadenylated 3’ends, along with the prokaryotic poly (A) tails are generally shorter and less mRNA molecules polyadenylated.
Mechanism of Polyadenylation
Polyadenylation process initiates in the nucleus of the eukaryotes which works in the RNA polymerase II, as a precursor mRNA, Where the 3’ part of the newly produced RNA and the polyadenylates are resultant of this cleavage process.
This process of cleavage is being catalyzed by the enzyme CPSF and it occurs 10-30 nucleotides downstream of its binding site.
This site mostly consists of a polyadenylation signal sequence AAUAA in the RNA, but the variants of it binds weaklier to the CPSF.
Further two other proteins are added specifically to the binding site of RNA; which are named as CstF and CFI, where the CstF binds to the GU region then downstream of the CPSFs site.
CFI recognizes at the third site on the RNA in a UGUAA sequence present in the mammals and recruits the CPSF IF THE AAUAA sequence is missing.
The polyadenylation signals sequence motif is recognized by the RNA cleavage complex which varies between the each of the eukaryotes.
Mostly the human polyadenylation sites contain the AAUAA sequence, but this sequence is less common in fungi and in plants.
RNA is cleaved before transcription termination because, CstF binds to the RNA polymerase II because the poorly understood mechanism which signals for the RNA polymerase II slip off out of the transcript.
Cleavage involves the protein CSF II though it is unknown how, the cleavage sites are associated with a polyadenylation signal it varies up to an average of 50 nucleotides.
During the cleaving of RNA, polyadenylation usually starts and it is catalyzed by the enzyme polyadenylate polymerase which builds the poly(A) tail which adds adenosine monophosphate units from the adenosine triphosphate to the RNA which cleaves off phosphate.
Another protein, PAB2 binds with the newly formed short poly (A) tail and it also increases the affinity of polyadenylate polymerase for the RNA.
When the poly(A) tail reaches an approximate of 250 nucleotides long it cannot bind with the CPSF and thus the polyadenylation stops determining the length of the poly(A) tail.
CPSF is in contact with the RNA polymerase II thus allowing it to rise a signal to the polymerase to end the process of transcription.
When the RNA polymerase II reaches a termination sequence, the transcription comes to an end with an signaling.
The polyadenylation mechanism is sometimes linked to the spliceosome physically which removes the introns from the RNAs.
Polyadenylation Citations
- Inference of the human polyadenylation code. Bioinformatics . 2018 Sep 1;34(17):2889-2898.
- RNA polyadenylation and its consequences in prokaryotes. Philos Trans R Soc Lond B Biol Sci . 2018 Nov 5;373(1762):20180166.
- Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet . 2013 Jul;14(7):496-506.
- Implications of polyadenylation in health and disease. Nucleus . 2014;5(6):508-19.
Share
Similar Post:
-

Transfer RNA (tRNA): Function, Definition, and Structure
Continue ReadingWhat is Transfer RNA (tRNA)?
Transfer RNA which is abbreviated as tRNA is also referred to s-RNA or Soluble RNA in older times.
It is considered as one of the adaptor molecules and it is composed of RNA containing about 76 to 90 nucleotides along the length which serves as the physical link between mRNA and the sequence of amino acid of proteins. t-RNA carries an amino acid for the synthesis of proteins in a cell to the ribosomes.
Complementation of the 3-nucleotide codon in the mRNA by the 3-nucleotide anticodon of the tRNA and results in synthesis of the proteins depending on the mRNA code.
tRNAs are also considered as the vital component for the process of translation which is one of the biological process in producing the new proteins following the genetic code.
Role of Transfer RNA (tRNA)
When the specific nucleotide sequences of the mRNA specify the amino acids, they are incorporated into the protein product which is known as gene further from that mRNA are transcribed.
The vital role of tRNA is to specify thy the sequences from the genetic code corresponding to that of the amino acid.
The mRNA encodes a protein with a series of codon, where each is recognized by a particular tRNA.
One end of tRNA matches a genetic codon with a three-nucleotide sequence called as anticodon.
The anticodon then forms 3 complementary base pairs with a codon in the mRNA during the biosynthesis of protein.
Where as on the other end the tRNA is covalently attached to the amino acid which corresponds to that of the anticodon sequence.
Each of the type of tRNA molecules are attached to their specific type of amino acid, so that each of the organism makes many types of amino acids.
There are many molecules of tRNA which have different anticodons and carry the similar amino acids.
The covalent attachment to the tRNA at the 3’ end is being catalyzed by an enzyme known as aminoacyl tRNA synthetases.
During the synthesis of protein, tRNAs attaches with the amnio acids which are then delivered to the ribosomes with the help of proteins which are known as elongation factors and it also aid in associating it with the ribosome, and synthesis a new polypeptide and translocate the ribosomes along with the mRNA.
If the anticodons of tRNAs matches the mRNA, another tRNA also bound to the ribosome and transfers the growing polypeptide chain from the 3’end of the amino acid attaches the 3’end of the newly delivered tRNA, and this reaction is being catalyzed by the ribosome.
Large number of the individual nucleotides in the tRNA may be modified chemically by methylation or deamidation.
This unusual bases at times affects the interaction with the tRNAs with ribosomes and sometimes it also occurs in anticodon to alter base pairing characters.
Structure of Transfer RNA (tRNA)
The transfer RNA structure can be categorized into primary and it is further transformed into secondary structure which looks similar to that of the clover leaf, and the tertiary structure is similar to that of the L-shaped 3D structure which allows it to fit in a P and A sites of the ribosome.
The clover structure transforms into the 3D L-shaped structure where the coaxial structures are present according to the stacking of the helices, and it is common to the tertiary structure of RNA.
The length of the arm as well as the loop in the t-RNA vary varies according to the species. The t-RNA species comprises of the following
5-terminal phosphate group.
Acceptor stem: It containing 7 to 9 base pairs and it is made by the 5’ terminal nucleotide with pairs with the 3’terminal nucleotide that contains CCA 3’ terminal group which is being attached to the amino acid. Usually, 3’terminal t-RNA structures are referred to as genomic tags. Acceptor stem contains non-Watson Crick base pairs.
CCA Tail: It is a sequence of cytosine-cytosine-Adenine which is present at the 3’end of the tRNA molecule. The amino acids are loaded with the tRNA by the enzyme aminoacyl tRNA synthetases, which forms aminoacyl-tRNA which is covalently bonded to the 3’hydroxyl group in the CCA tail. This sequence is very much important for the recognition of the tRNA by certain enzymes and it is critical during translation. Where as in prokaryotes CCA sequence is transcribed into some tRNA sequences. In most prokaryotes tRNAs and the eukaryotic tRNAs the sequence of CCA is added during the process and therefore it does not appear in the tRNA gene.
D-Arm: It is made up of 4 to 6 base pair stem which ends in a loop and often contains dihydrouridine.
Anticodon Arm: It is made up of five base pairs and looks like a stem and ends in a loop which contains anticodon. In tRNA, 5’to 3’ end contains anticodon. But it is present in the reverse order, therefore, 3’to 5’ directionality is needed to read the mRNA from 5’to 3’end.
T-Arm: The T-arm in the transfer RNA is made up of about five base pairs which contains the sequence of TψC, where ψ refers to pseudo uridine which is a modified form of uridine.
Bases in the tRNA are modified by the process of methylation, which occur in several positions throughout the tRNA. The first base of the anticodon is wobble in position and it is sometimes modified as inosine- derived from adenine; queosine which is derived from Guanine; 5-methylaminomethyl-2-thiouridine is derived from the uracil and Uridine – – oxyacetic acid which is derived from uracil or as lysidine which is derived from Cytosine.
Transfer RNA (tRNA) Citations
Share
Similar Post:
-
Ribosomal RNA (rRNA): Function, Definition, and Structure
Continue ReadingWhat is Ribosomal RNA (rRNA)?
Ribosomal ribonucleic acid is one of the types of non-coding RNA which is one of the primary components in the ribosomes, and it is also very important for all the cells.
rRNA is also known as ribozyme that carries out a protein synthesis in the cell organelle ribosomes, this ribosomal RNA is transcribed from the ribosomal DNA and then it is bound to the ribosomal proteins and forms small and large subunits of the ribosomes.
rRNA is considered as a physical and a mechanical factor for the ribosome as it forces the transfer of RNA and the mRNA which further process and translates into proteins.
Ribosomal RNA is the predominant form of RNA found in most of the cells and it also makes about eighty percentage of the cellular RNA despites never been translated into the proteins by itself.
Ribosomes are made up of approximately 60% of the rRNA and 40% of the other ribosomal protein mass.
Ribosomal RNA (rRNA) molecule in the cells that forms part of the proteins synthesizing organelle which is known ribosome which exported to the cytoplasm of the cells which helps in translating the information in the messenger RNA as protein.
Features of Ribosomal RNA (rRNA)
Ribosomal ribo nucleic acid which is generally referred to as rRNA is the component of the ribosomes, the molecular machines which catalyzes the synthesis of proteins.
rRNA constitutes about six percentage of the ribosomes by weight and are very crucial in performing their functions, such as binding to the mRNA and recruiting the tRNA in catalyzing the formation of the peptide bond in between the two amino acids.
Though the structure od a ribosome is determined by a three-dimensional shape in its rRNA core. Proteins which are present in the ribosomes serves in stabling the structure by interacting with its core.
Ribosomal RNA is transcribed within the nucleus and particularly in the nucleoli which is present inside the nucleus.
They are usually dense, spherical shape which is formed around the loci of the genome and codes for rRNA. Nucleoli are also stable in biogenies of the ribosomes by sequestration of ribosomal proteins.
Discovery of Ribosomal RNA (rRNA)
The Ribosomal RNA was first discovered during the fractionation of the cell experiments which invests the role of RNA viruses which causes cancer.
Fractionation is a method where cell membranes carefully removed in a selective manner by keeping the cellular organelles function intact.
This type of homogenized cytoplasm is further centrifuged at an increasing speed so that the organelles are separated according to the density.
The initial experiments which reveal the presence of rRNA are extracted as a fracted which was thought to represent a new organelle, which is sub-cellular known as microsome – specialized in protein synthesis.
Further it was observed that if the presence of the ribosomes in the endoplasmic reticulum helps in detecting the samples of RNA.
As ribosomal subunits and the rRNA were detected initially by differential centrifugation they are still characterized by their rate of sedimentation with the help of Svedberg coefficient.
As these are not the direct measures of the molecular weight the coefficients are not added directly.
Considering an example of prokaryotic ribosomes where it has a larger subunits 50S and a smaller subunit of 30S comes together as a complex and has a Svedberg co efficient of 70S.
Types of Ribosomal RNA (rRNA)
Both prokaryotic and eukaryotic ribosomes are made up of larger and a smaller sub unit and these units come together during the translation of mRNA.
Where as the smaller sub unit of the RNA is made up of 1500 nucleotides in length having a Svedberg coefficient of 16S. Along with the ribosomal proteins the smaller sub units have a sedimentation rate of about 30S, which is paired with the larger sub unit that has two molecules of RNA which has nearly 3000 nucleotides and has a sedimentation rate of about 23S and a short sequence of about 120 nucleotides and has a rate of about 5S.
The RNA molecules are accompanied by the proteins which gives rise to larger sub units of 50S.
The eukaryotic ribosomes are made up of about 60S and a 40S sub unit. It has two short rRNA molecules that are less than two hundred nucleotides in length and the two molecules of RNA are much longer.
In addition to all these eukaryotic ribosomes also have rRNA in mitochondria and the chloroplasts.
Ribosomes are also associated with the endoplasmic reticulum or it is present in a free-floating complex of the cytoplasm.
Function of Ribosomal RNA (rRNA)
One of the primary functions of the ribosomal RNA is the synthesis of protein, and in binding to the messenger RNA and the transfer RNA and in ensuring the codon sequence of the messenger RNA, and it is translated accurately into an amino acid sequence in the form of proteins.
In order to achieve this, rRNA has a complex three-dimensional structure which involves an internal loop and the helices and creates a specific site inside the ribosomes known as A, P and E sites.
The P site is especially for binding the growing polypeptide and the site A anchors an incoming transfer RNA which is charged with amino acid.
After the formation of a peptide bond, the transfer RNA binds shortly along with the E- site before it is leaving the ribosome.
In addition to this rRNA has also has specific sites to bind with the few ribosomal proteins and it also carefully analysis the exact residues that is present in both the RNA and the protein. Ribosomal RNAs can also be expressed in cells of the extant species.
The sequence of the core catalytic sites is highly conserved by making rRNA as a superficial tool for studying the taxonomy and the phylogenetics.
It has also been found the difference in the rate of evolution of residues on the surfaces which are interior to that of the rRNA and the nucleotides which are involved in the catalytic activity, which informs formation of a peptide bond which are appeared to have predated the appearance of life on earth.
Many of the antibiotics mostly targets the rRNA of the prokaryotes and in recent times it has been found that the binding sites for the antibiotics like streptomycin and tetracycline have been found. It has also been observed that the antibiotic resistance forms point mutations in the binding sites.
Ribosomal RNA (rRNA) Citations
Share
Similar Post:
-
Cistron: Definition, Function, and Example
Continue ReadingCistron, Chromosome, and Gene
In the earlier period it was said that a gene is the smallest unit which is present on the chromosome, and it is also considered as the structural and functional unit of a genome, and it is said that gene is an important molecule in causing mutations and which is also important in recombination.
It has also been believed that crossing over happens in between the genes and it does not occur between the same genes. These concepts were found in the experiments done in early stages.
But on later researches it has been clear that crossing over occurs by the breakage and reunion of the DNA molecules, when the genes are closely spaced such as in case f multiple alleles it results in rare frequency of intragenic recombination, so that a very large test cross progeny is required for its detection.
Occurring of mutant alleles gave an insight into the composition of the gene. Such alleles in a gene are separated by a small distance within themselves and are related functions.
Therefore, a unit of gene is a cistron. Generally, a genetic test is used to define the cistrons.
What is Cistron?
The term cistron was first coined even before the advances that are being made in biology, by the scientist named Seymour Benzer in the article the elementary unit if hereditary cistron is shortly defined as the alternative term of gene.
The term cistron is often used to emphasize the genes that exhibit a specific behavior in cis-trans set, with distinct loci within their genome as cistron.
Cistron is generally a segment of DNA, which is equivalent to a gene. It is also considered as the smallest unit of the genetic material that codes for single polypeptide and their functions by acting as a transmitter for transmitting genetic information.
Hence it is said that gene consists of more than a one cistron.
Mutation and Cistron
To understand the function of cistron, considering an example of mutation, Mutation that is taking part due to the position of the chromosomes. Which is responsible for the changes caused in a recessive trait in the diploid organism.
We say, Mutation is recessive until both chromosomes of a pair have mutation, in a particular trait, will not express any disturbances or defects.
On the other hand, if mutation takes place in any other position B which is responsible for the same recessive trait; then it is considered that both the positions A and B are in the same cistron.
We can take a hypothetical gene containing two alleles which is mutant in condition and they are assumed as α1 and α2.
When these two alleles are present in different numbers in a pair of homologous chromosomes, the mutant phenotype is produced.
The alleles that are present is in opposite positions on the two chromosomes which are said be trans arrangement and they are also non-complementing because they produce a mutant phenotype that is visible.
If both the alleles are present on the same chromosome, then it is said to be as Cis- arrangement which produces a wild phenotype.
Let us consider a third allele termed as α3 which is present on s trans arrangement with the α1 allele in the paired chromosome, it also results in the formation of a wild phenotype.
In such cases it is said that the alleles α1 and α3 are considered to be complementary to each other.
Hence it is said that when 2 mutations are present in the trans position, it produces a mutant phenotype and it is also said to be member of the same functional unit, which is said to be the cistron.
But in other case, where the two mutations are present in the trans position complementing each other then it is said that they belong to different cistrons.
From the concept of the cistron it is said that cistron is dependent on the position of cis- trans effect.
The scientist, Lewis from his work in 1951 devised the effect of Cis- trans test for the complementarity between two alleles.
During essence, it consists of comparing the phenotypes which are produced between two mutations, when they are present in the Cis-trans configuration.
When considering the terms of complementation, Cistron is used instead in the place of gene. The gene is a functional unit in a sequence of nucleotides in the molecule of DNA which codes for one of the polypeptide chains.
This genetic complementation is also applicable for haploid organisms like that of Neurospora. Where as in higher organisms, the complementation can be extensively studied in insects like Drosophila.
But in some cases, it is difficult to identify and understand that the functional product of gene is a protein which is unrelated at times.
An abnormal eye condition in Drosophila is called as lozenge which was used by the researcher Green and Green in the year 1949 for mapping the alleles on the locus.
This sequence of mutant sites was determined by the frequency pf intragenic recombination that is occurring because of the rare frequency, which is required for detecting the large test cross progeny.
They performed testcrosses of the heterozygous females for the different lozenge alleles and they obtained a single lozenge gene on a linear map.
These intergenic recombination of the alleles are said to be as fine structure mapping,
Cistron Citations
Share
Similar Post: